[English] 日本語
 Yorodumi
Yorodumi- EMDB-14750: Delta SARS-CoV-2 spike protein in complex with REGN10987 Fab homo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
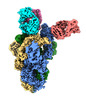
| Title | Delta SARS-CoV-2 spike protein in complex with REGN10987 Fab homologue. | |||||||||
 Map data Map data | Composite map created with locally sharpened RBD-FAB structure after local refinement and a C1 map of the complex. See a C1 map with the half-maps attached. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / REGN10987 / Fab / cryo-EM / S-protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
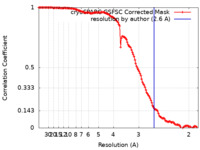
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Pichkur EB / Lyukmanova EN / Shenkarev ZO | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Delta SARS-CoV-2 spike protein in complex with REGN10987 Fab homologue. Authors: Pichkur EB / Lyukmanova EN / Shenkarev ZO | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14750.map.gz emd_14750.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14750-v30.xml emd-14750-v30.xml emd-14750.xml emd-14750.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14750_fsc.xml emd_14750_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14750.png emd_14750.png | 119.2 KB | ||
| Masks |  emd_14750_msk_1.map emd_14750_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14750.cif.gz emd-14750.cif.gz | 6.5 KB | ||
| Others |  emd_14750_additional_1.map.gz emd_14750_additional_1.map.gz emd_14750_half_map_1.map.gz emd_14750_half_map_1.map.gz emd_14750_half_map_2.map.gz emd_14750_half_map_2.map.gz | 204.3 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14750 http://ftp.pdbj.org/pub/emdb/structures/EMD-14750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14750 | HTTPS FTP |
-Related structure data
| Related structure data |  7zjlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14750.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14750.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map created with locally sharpened RBD-FAB structure after local refinement and a C1 map of the complex. See a C1 map with the half-maps attached. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.929 Å | ||||||||||||||||||||||||||||||||||||
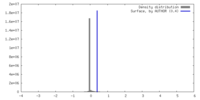
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14750_msk_1.map emd_14750_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
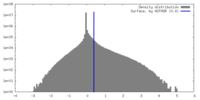
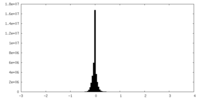
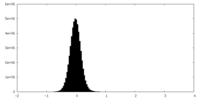
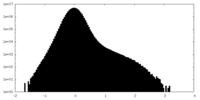
| Density Histograms |
-Additional map: Sharpened C1 map
| File | emd_14750_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened C1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
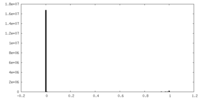
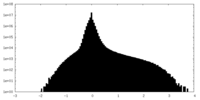
| Density Histograms |
-Half map: #1
| File | emd_14750_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
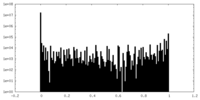
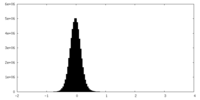
| Density Histograms |
-Half map: #2
| File | emd_14750_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
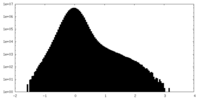
| Density Histograms |
- Sample components
Sample components
-Entire : Delta SARS-CoV-2 S-protein + REGN10987 Fab homologue
| Entire | Name: Delta SARS-CoV-2 S-protein + REGN10987 Fab homologue |
|---|---|
| Components |
|
-Supramolecule #1: Delta SARS-CoV-2 S-protein + REGN10987 Fab homologue
| Supramolecule | Name: Delta SARS-CoV-2 S-protein + REGN10987 Fab homologue / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Delta SARS-CoV-2 S-protein
| Supramolecule | Name: Delta SARS-CoV-2 S-protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: REGN10987 Fab homologue
| Supramolecule | Name: REGN10987 Fab homologue / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 125.147805 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CVNLRTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV YYHKNNKSWM ESGVYSSANN CTFEYVSQPF L MDLEGKQG ...String: CVNLRTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV YYHKNNKSWM ESGVYSSANN CTFEYVSQPF L MDLEGKQG NFKNLREFVF KNIDGYFKIY SKHTPINLVR DLPQGFSALE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SS GWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLC PFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTG KIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY RYRLFRKSNL KPFERDISTE IYQAGSKPCN GVEGFNCYFP LQSYG FQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAV RDP QTLEILDITP CSFGGVSVIT PGTNTSNQVA VLYQGVNCTE VPVAIHADQL TPTWRVYSTG SNVFQTRAGC LIGAEHV NN SYECDIPIGA GICASYQTQT NSPRRARSVA SQSIIAYTMS LGAENSVAYS NNSIAIPTNF TISVTTEILP VSMTKTSV D CTMYICGDST ECSNLLLQYG SFCTQLNRAL TGIAVEQDKN TQEVFAQVKQ IYKTPPIKDF GGFNFSQILP DPSKPSKRS FIEDLLFNKV TLADAGFIKQ YGDCLGDIAA RDLICAQKFN GLTVLPPLLT DEMIAQYTSA LLAGTITSGW TFGAGAALQI PFAMQMAYR FNGIGVTQNV LYENQKLIAN QFNSAIGKIQ DSLSSTASAL GKLQNVVNQN AQALNTLVKQ LSSNFGAISS V LNDILSRL DKVEAEVQID RLITGRLQSL QTYVTQQLIR AAEIRASANL AATKMSECVL GQSKRVDFCG KGYHLMSFPQ SA PHGVVFL HVTYVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IITTDNTFVS GNCDVVIGIV NNT VYDPLQ PELD UniProtKB: Spike glycoprotein |
-Macromolecule #2: REGN10987 Fab homologue (Light chain)
| Macromolecule | Name: REGN10987 Fab homologue (Light chain) / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.334691 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCTGTSSDVG GYNYVSWYQQ HPGKAPKLMI YDVSKRPSGV SNRFSGSKSG NTASLTISGL QSEDEADYY CNSLTSISTW VFGGGTKLTV LGRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT ...String: QSALTQPASV SGSPGQSITI SCTGTSSDVG GYNYVSWYQQ HPGKAPKLMI YDVSKRPSGV SNRFSGSKSG NTASLTISGL QSEDEADYY CNSLTSISTW VFGGGTKLTV LGRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #3: REGN10987 Fab homologue (Heavy chain)
| Macromolecule | Name: REGN10987 Fab homologue (Heavy chain) / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.792617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS NYAMYWVRQA PGKGLEWVAV ISYDGSNKYY ADSVKGRFTI SRDNSKNTLY LQMNSLRTE DTAVYYCASG SDYGDYLLVY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS ...String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS NYAMYWVRQA PGKGLEWVAV ISYDGSNKYY ADSVKGRFTI SRDNSKNTLY LQMNSLRTE DTAVYYCASG SDYGDYLLVY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: 20 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)