+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
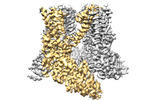
| Title | TRPV2-C16+Pro-2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Temperature sensor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgrowth cone membrane / response to temperature stimulus / positive regulation of calcium ion import / axonal growth cone / positive regulation of axon extension / calcium channel activity / positive regulation of cold-induced thermogenesis / cell body / cell surface Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.39 Å | |||||||||
 Authors Authors | Zhang L / Gourdon P / Zygmunt PM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function. Authors: Liying Zhang / Charlotte Simonsen / Lucie Zimova / Kaituo Wang / Lavanya Moparthi / Rachelle Gaudet / Maria Ekoff / Gunnar Nilsson / Ute A Hellmich / Viktorie Vlachova / Pontus Gourdon / Peter M Zygmunt /      Abstract: TRPV2 is a ligand-operated temperature sensor with poorly defined pharmacology. Here, we combine calcium imaging and patch-clamp electrophysiology with cryo-electron microscopy (cryo-EM) to explore ...TRPV2 is a ligand-operated temperature sensor with poorly defined pharmacology. Here, we combine calcium imaging and patch-clamp electrophysiology with cryo-electron microscopy (cryo-EM) to explore how TRPV2 activity is modulated by the phytocannabinoid Δ-tetrahydrocannabiorcol (C16) and by probenecid. C16 and probenecid act in concert to stimulate TRPV2 responses including histamine release from rat and human mast cells. Each ligand causes distinct conformational changes in TRPV2 as revealed by cryo-EM. Although the binding for probenecid remains elusive, C16 associates within the vanilloid pocket. As such, the C16 binding location is distinct from that of cannabidiol, partially overlapping with the binding site of the TRPV2 inhibitor piperlongumine. Taken together, we discover a new cannabinoid binding site in TRPV2 that is under the influence of allosteric control by probenecid. This molecular insight into ligand modulation enhances our understanding of TRPV2 in normal and pathophysiology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14748.map.gz emd_14748.map.gz | 91.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14748-v30.xml emd-14748-v30.xml emd-14748.xml emd-14748.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14748.png emd_14748.png | 100.8 KB | ||
| Filedesc metadata |  emd-14748.cif.gz emd-14748.cif.gz | 6.2 KB | ||
| Others |  emd_14748_half_map_1.map.gz emd_14748_half_map_1.map.gz emd_14748_half_map_2.map.gz emd_14748_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14748 http://ftp.pdbj.org/pub/emdb/structures/EMD-14748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14748 | HTTPS FTP |
-Validation report
| Summary document |  emd_14748_validation.pdf.gz emd_14748_validation.pdf.gz | 948.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14748_full_validation.pdf.gz emd_14748_full_validation.pdf.gz | 948.4 KB | Display | |
| Data in XML |  emd_14748_validation.xml.gz emd_14748_validation.xml.gz | 15.1 KB | Display | |
| Data in CIF |  emd_14748_validation.cif.gz emd_14748_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14748 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14748 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14748 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14748 | HTTPS FTP |
-Related structure data
| Related structure data |  7zjhMC  7zjdC  7zjeC  7zjgC  7zjiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14748.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14748.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8464 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14748_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14748_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRPV2-C16+Pro-2
| Entire | Name: TRPV2-C16+Pro-2 |
|---|---|
| Components |
|
-Supramolecule #1: TRPV2-C16+Pro-2
| Supramolecule | Name: TRPV2-C16+Pro-2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 2
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 2 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.426359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSASSPPAF RLETSDGDEE GNAEVNKGKQ EPPPMESPFQ REDRNSSPQI KVNLNFIKRP PKNTSAPSQQ EPDRFDRDRL FSVVSRGVP EELTGLLEYL RWNSKYLTDS AYTEGSTGKT CLMKAVLNLQ DGVNACIMPL LQIDKDSGNP KPLVNAQCID E FYQGHSAL ...String: MTSASSPPAF RLETSDGDEE GNAEVNKGKQ EPPPMESPFQ REDRNSSPQI KVNLNFIKRP PKNTSAPSQQ EPDRFDRDRL FSVVSRGVP EELTGLLEYL RWNSKYLTDS AYTEGSTGKT CLMKAVLNLQ DGVNACIMPL LQIDKDSGNP KPLVNAQCID E FYQGHSAL HIAIEKRSLQ CVKLLVENGA DVHLRACGRF FQKHQGTCFY FGELPLSLAA CTKQWDVVTY LLENPHQPAS LE ATDSLGN TVLHALVMIA DNSPENSALV IHMYDGLLQM GARLCPTVQL EEISNHQGLT PLKLAAKEGK IEIFRHILQR EFS GPYQPL SRKFTEWCYG PVRVSLYDLS SVDSWEKNSV LEIIAFHCKS PNRHRMVVLE PLNKLLQEKW DRLVSRFFFN FACY LVYMF IFTVVAYHQP SLDQPAIPSS KATFGESMLL LGHILILLGG IYLLLGQLWY FWRRRLFIWI SFMDSYFEIL FLLQA LLTV LSQVLRFMET EWYLPLLVLS LVLGWLNLLY YTRGFQHTGI YSVMIQKVIL RDLLRFLLVY LVFLFGFAVA LVSLSR EAR SPKAPEDSSS TVTEQPTVGQ EEEPAPYRSI LDASLELFKF TIGMGELAFQ EQLRFRGVVL LLLLAYVLLT YVLLLNM LI ALMSETVNHV ADNSWSIWKL QKAISVLEME NGYWWCRRKK HREGRLLKVG TRGDGTPDER WCFRVEEVNW AAWEKTLP T LSEDPSGPGI TGNKKNPTSK PGKNSASEED HLPLQVLQSP AAAGLVPRGS VAAAVSKGEE LFTGVVPILV ELDGDVNGH KFSVSGEGEG DATYGKLTLK FICTTGKLPV PWPTLVTTLT YGVQCFSRYP DHMKQHDFFK SAMPEGYVQE RTIFFKDDGN YKTRAEVKF EGDTLVNRIE LKGIDFKEDG NILGHKLEYN YNSHNVYIMA DKQKNGIKVN FKIRHNIEDG SVQLADHYQQ N TPIGDGPV LLPDNHYLST QSKLSKDPNE KRDHMVLLEF VTAAGITLGM DELYKSGLRS HHHHHHHH UniProtKB: Transient receptor potential cation channel, subfamily V, member 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)