+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | X-31 Hemagglutinin Precursor HA0 at pH 4.8 | |||||||||
 Map data Map data | Sharpened map of the X-31 influenza hemagglutinin precursor HA0 at low pH. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus envelope / receptor binding / membrane fusion / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / virion component / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/Aichi/2/1968(H3N2)) / Influenza A virus (A/Aichi/2/1968(H3N2)) /  Tequatrovirus T4 Tequatrovirus T4 | |||||||||
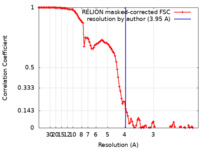
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Garcia-Moro E / Rosenthal PB | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Reversible structural changes in the influenza hemagglutinin precursor at membrane fusion pH. Authors: Eva Garcia-Moro / Jie Zhang / Lesley J Calder / Nick R Brown / Steven J Gamblin / John J Skehel / Peter B Rosenthal /  Abstract: The subunits of the influenza hemagglutinin (HA) trimer are synthesized as single-chain precursors (HA0s) that are proteolytically cleaved into the disulfide-linked polypeptides HA1 and HA2. Cleavage ...The subunits of the influenza hemagglutinin (HA) trimer are synthesized as single-chain precursors (HA0s) that are proteolytically cleaved into the disulfide-linked polypeptides HA1 and HA2. Cleavage is required for activation of membrane fusion at low pH, which occurs at the beginning of infection following transfer of cell-surface-bound viruses into endosomes. Activation results in extensive changes in the conformation of cleaved HA. To establish the overall contribution of cleavage to the mechanism of HA-mediated membrane fusion, we used cryogenic electron microscopy (cryo-EM) to directly image HA0 at neutral and low pH. We found extensive pH-induced structural changes, some of which were similar to those described for intermediates in the refolding of cleaved HA at low pH. They involve a partial extension of the long central coiled coil formed by melting of the preexisting secondary structure, threading it between the membrane-distal domains, and subsequent refolding as extended helices. The fusion peptide, covalently linked at its N terminus, adopts an amphipathic helical conformation over part of its length and is repositioned and packed against a complementary surface groove of conserved residues. Furthermore, and in contrast to cleaved HA, the changes in HA0 structure at low pH are reversible on reincubation at neutral pH. We discuss the implications of covalently restricted HA0 refolding for the cleaved HA conformational changes that mediate membrane fusion and for the action of antiviral drug candidates and cross-reactive anti-HA antibodies that can block influenza infectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14743.map.gz emd_14743.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14743-v30.xml emd-14743-v30.xml emd-14743.xml emd-14743.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14743_fsc.xml emd_14743_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14743.png emd_14743.png | 74 KB | ||
| Masks |  emd_14743_msk_1.map emd_14743_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14743.cif.gz emd-14743.cif.gz | 7.1 KB | ||
| Others |  emd_14743_additional_1.map.gz emd_14743_additional_1.map.gz emd_14743_half_map_1.map.gz emd_14743_half_map_1.map.gz emd_14743_half_map_2.map.gz emd_14743_half_map_2.map.gz | 91.3 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14743 http://ftp.pdbj.org/pub/emdb/structures/EMD-14743 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14743 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14743 | HTTPS FTP |
-Related structure data
| Related structure data |  7zj7MC  7zj6C  7zj8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14743.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14743.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of the X-31 influenza hemagglutinin precursor HA0 at low pH. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
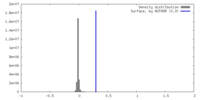
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14743_msk_1.map emd_14743_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
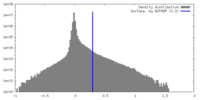
| Density Histograms |
-Additional map: Map processed with the deepEMhancer software, used to...
| File | emd_14743_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map processed with the deepEMhancer software, used to aid model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
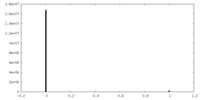
| Density Histograms |
-Half map: Half map 2.
| File | emd_14743_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
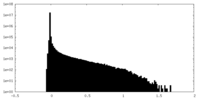
| Density Histograms |
-Half map: Half map 1.
| File | emd_14743_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : X-31 Hemagglutinin Precursor (HA0)
| Entire | Name: X-31 Hemagglutinin Precursor (HA0) |
|---|---|
| Components |
|
-Supramolecule #1: X-31 Hemagglutinin Precursor (HA0)
| Supramolecule | Name: X-31 Hemagglutinin Precursor (HA0) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Uncleaved precursor form of the X-31 influenza hemagglutinin with the T4 fibritin foldon attached to the C-terminus. |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Aichi/2/1968(H3N2)) Influenza A virus (A/Aichi/2/1968(H3N2)) |
-Macromolecule #1: Hemagglutinin,Fibritin
| Macromolecule | Name: Hemagglutinin,Fibritin / type: protein_or_peptide / ID: 1 Details: T4 fibritin foldon attached to the C-terminal of HA0 as a trimerization domain. C-terminal His8-tag for protein purification.,T4 fibritin foldon attached to the C-terminal of HA0 as a ...Details: T4 fibritin foldon attached to the C-terminal of HA0 as a trimerization domain. C-terminal His8-tag for protein purification.,T4 fibritin foldon attached to the C-terminal of HA0 as a trimerization domain. C-terminal His8-tag for protein purification.,T4 fibritin foldon attached to the C-terminal of HA0 as a trimerization domain. C-terminal His8-tag for protein purification.,T4 fibritin foldon attached to the C-terminal of HA0 as a trimerization domain. C-terminal His8-tag for protein purification. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 62.027055 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QDLPGNDNST ATLCLGHHAV PNGTLVKTIT DDQIEVTNAT ELVQSSSTGK ICNNPHRILD GIDCTLIDAL LGDPHCDVFQ NETWDLFVE RSKAFSNCYP YDVPDYASLR SLVASSGTLE FITEGFTWTG VTQNGGSNAC KRGPGSGFFS RLNWLTKSGS T YPVLNVTM ...String: QDLPGNDNST ATLCLGHHAV PNGTLVKTIT DDQIEVTNAT ELVQSSSTGK ICNNPHRILD GIDCTLIDAL LGDPHCDVFQ NETWDLFVE RSKAFSNCYP YDVPDYASLR SLVASSGTLE FITEGFTWTG VTQNGGSNAC KRGPGSGFFS RLNWLTKSGS T YPVLNVTM PNNDNFDKLY IWGIHHPSTN QEQTSLYVQA SGRVTVSTRR SQQTIIPNIG SRPWVRGLSS RISIYWTIVK PG DVLVINS NGNLIAPRGY FKMRTGKSSI MRSDAPIDTC ISECITPNGS IPNDKPFQNV NKITYGACPK YVKQNTLKLA TGM RNVPEK QTRGLFGAIA GFIENGWEGM IDGWYGFRHQ NSEGTGQAAD LKSTQAAIDQ INGKLNRVIE KTNEKFHQIE KEFS EVEGR IQDLEKYVED TKIDLWSYNA ELLVALENQH TIDLTDSEMN KLFEKTRRQL RENAEEMGNG CFKIYHKCDN ACIES IRNG TYDHDVYRDE ALNNRFQIKG GGRENLYFQG GGGSGYIPEA PRDGQAYVRK DGEWVLLSTF LGHHHHHHHH UniProtKB: Hemagglutinin, Fibritin |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 4.8 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 4s blot. |
| Details | The sample protein tended to aggregate and bind to the carbon film. 0.1% b-octyl glucoside was added. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-32 / Number grids imaged: 2 / Number real images: 33977 / Average exposure time: 8.0 sec. / Average electron dose: 41.15 e/Å2 Details: 15696 images on session 1, 18281 images on session 2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)