+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S-layer Deinoxanthin Binding Complex, C3 symmetry | |||||||||
 Map data Map data | Complex C3 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PROTEIN / STRUCTURAL PROTEIN / S-layer / deinococcus radiodurans / DR_2577 / S-layer Deinoxanthin Binding Complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationporin activity / pore complex / monoatomic ion transport / cell outer membrane / lipid binding Similarity search - Function | |||||||||
| Biological species |  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.54 Å | |||||||||
 Authors Authors | Farci D / Piano D | |||||||||
| Funding support |  Poland, 2 items Poland, 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions. Authors: Domenica Farci / Patrycja Haniewicz / Daniele de Sanctis / Luca Iesu / Sami Kereïche / Mathias Winterhalter / Dario Piano /       Abstract: The radiation-resistant bacterium Deinococcus radiodurans is known as the world's toughest bacterium. The S-layer of D. radiodurans, consisting of several proteins on the surface of the cellular ...The radiation-resistant bacterium Deinococcus radiodurans is known as the world's toughest bacterium. The S-layer of D. radiodurans, consisting of several proteins on the surface of the cellular envelope and intimately associated with the outer membrane, has therefore been useful as a model for structural and functional studies. Its main proteinaceous unit, the S-layer deinoxanthin-binding complex (SDBC), is a hetero-oligomeric assembly known to contribute to the resistance against environmental stress and have porin functional features; however, its precise structure is unknown. Here, we resolved the structure of the SDBC at ∼2.5 Å resolution by cryo-EM and assigned the sequence of its main subunit, the protein DR_2577. This structure is characterized by a pore region, a massive β-barrel organization, a stalk region consisting of a trimeric coiled coil, and a collar region at the base of the stalk. We show that each monomer binds three Cu ions and one Fe ion and retains one deinoxanthin molecule and two phosphoglycolipids, all exclusive to D. radiodurans. Finally, electrophysiological characterization of the SDBC shows that it exhibits transport properties with several amino acids. Taken together, these results highlight the SDBC as a robust structure displaying both protection and sieving functions that facilitates exchanges with the environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14715.map.gz emd_14715.map.gz | 530.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14715-v30.xml emd-14715-v30.xml emd-14715.xml emd-14715.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14715_fsc.xml emd_14715_fsc.xml | 18.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14715.png emd_14715.png | 110.7 KB | ||
| Filedesc metadata |  emd-14715.cif.gz emd-14715.cif.gz | 6.8 KB | ||
| Others |  emd_14715_half_map_1.map.gz emd_14715_half_map_1.map.gz emd_14715_half_map_2.map.gz emd_14715_half_map_2.map.gz | 465.8 MB 477.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14715 http://ftp.pdbj.org/pub/emdb/structures/EMD-14715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14715 | HTTPS FTP |
-Related structure data
| Related structure data |  7zgyMC  8acaM  8acqM  7zgxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14715.map.gz / Format: CCP4 / Size: 561.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14715.map.gz / Format: CCP4 / Size: 561.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex C3 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.818 Å | ||||||||||||||||||||||||||||||||||||
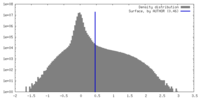
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Complex C3 Half map A
| File | emd_14715_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex C3 Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
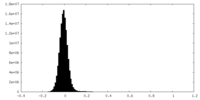
| Density Histograms |
-Half map: Complex C3 Half map B
| File | emd_14715_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex C3 Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
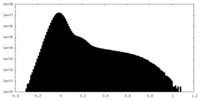
| Density Histograms |
- Sample components
Sample components
-Entire : S-layer Deinoxanthin Binding Complex, C3 Symmetry
| Entire | Name: S-layer Deinoxanthin Binding Complex, C3 Symmetry |
|---|---|
| Components |
|
-Supramolecule #1: S-layer Deinoxanthin Binding Complex, C3 Symmetry
| Supramolecule | Name: S-layer Deinoxanthin Binding Complex, C3 Symmetry / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant) |
| Molecular weight | Theoretical: 800 KDa |
-Macromolecule #1: S-layer protein SlpA
| Macromolecule | Name: S-layer protein SlpA / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans R1 (radioresistant) Deinococcus radiodurans R1 (radioresistant)Strain: ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422 |
| Molecular weight | Theoretical: 123.835367 KDa |
| Sequence | String: MKKSLIALTT ALSFGLAAAQ TAAPVSAPQV PALTDVPAGH WAKDAIDRLV SRGVILGYPD GTFRGTQNLT RYEAAIIIAR LLDQMRDGE TPAGMTAEDM TALQNAIQEL AADLAALGVR VSDLEANAVS KDDFARLEAR IEEVAAAGGE QGATEALQGQ I DDLTARVD ...String: MKKSLIALTT ALSFGLAAAQ TAAPVSAPQV PALTDVPAGH WAKDAIDRLV SRGVILGYPD GTFRGTQNLT RYEAAIIIAR LLDQMRDGE TPAGMTAEDM TALQNAIQEL AADLAALGVR VSDLEANAVS KDDFARLEAR IEEVAAAGGE QGATEALQGQ I DDLTARVD EYDALRADVD DNASSIAALN DLTVLLNQDI LDLQDRVSAV EAAQADFVQR SDFDALGGRV TTVETRVETV NN SLTGRIA ALERNAFSVK PSLTIGYSVS RTSRNFDVDR LFPLNADGTV ANNAFTSGGI DTDTGAQRRD FGDFGNASDP VVA GAAGLY GFADGVSYTV YFTDGSTATF DGLNPADYKV PTGKVIDTTK GRNGFGFNNL ARYKEGSTDI GISLGFDTSG QFSQ VTSGT GGSLFSTAGR LQVNQIDLNF GLVTGLPSDA YVDTNGNGKK DDGEATGRGT YLGSGGTAAI LRDPAGNVYR PVFFR FKNA TTQFSVGNNP VIVTLGQQQK FYFSDYVFDN NYDGRGDGFT VTVDGSNVPV IGAWKPQIKG VYGSRSGLDG TAEAGY GVY YRGVRAQITP VGTLTAGIHY AQEGRDMFGA AQNTTSTPSD VTTYGADLHG KAFGVELHSE YATSRVRPNT ANAAVQT SN AFYARVATRK DNLAFDLNTP AAKFGNDTFG VSLYDLNYRK IDAGYNNVAG ISEYGYGSYS RTSAQNIAYN PDTGVTAP F ANLDRQAYTD ANNDGTSDRN ADGTVVATNT KIGQMGFGVK AAANLGPVAI GGYYDTSTGA NGDNANRMTE AGGSAKVAY SIFSLRGTYN TLDSNRPQIY RDAAGTQIIG DAKVRRYAVQ ADVTPGLGLF VGAYYRDVNV NGVRSTTDRG LLGRGYLASS FEPGVGNNA YRTGLRCADN NFGTGTRDID GVGGVLNPAV NLDQSRTATC FTSYGVEAGH AGDNANALVK DLFFRVGYSR V YVPTTATA TTGDFSGSVT YGDARYDRKV GVANVRLAGS FSTTNTQLDS RPAGTRGAVG LIVRTDPLEN VPFRPQFNGQ VG YYTADNR VAAGNYNANA TKYGAGVVLN DFLLPQTKIG VRYDGYMAQN RQYTPFDGDG TQGYFSDANN NRRTNLNGVY VEG AYQDLI FSYGTYTLSQ KDLNGVEYGS GINNGQPARG QTFKISYKVN F UniProtKB: Outer membrane protein SlpA |
-Macromolecule #2: (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,...
| Macromolecule | Name: (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol type: ligand / ID: 2 / Number of copies: 3 / Formula: JPI |
|---|---|
| Molecular weight | Theoretical: 609.061 Da |
| Chemical component information |  ChemComp-JPI: |
-Macromolecule #3: [(2~{S})-2-acetyloxy-3-[[(2~{S})-3-[(2~{R},3~{S},4~{S},5~{S},6~{S...
| Macromolecule | Name: [(2~{S})-2-acetyloxy-3-[[(2~{S})-3-[(2~{R},3~{S},4~{S},5~{S},6~{S})-6-(hydroxymethyl)-3-(octadecanoylamino)-4,5-bis(oxidanyl)oxan-2-yl]oxy-1-oxidanylidene-1-(pentylamino)propan-2-yl]oxy- ...Name: [(2~{S})-2-acetyloxy-3-[[(2~{S})-3-[(2~{R},3~{S},4~{S},5~{S},6~{S})-6-(hydroxymethyl)-3-(octadecanoylamino)-4,5-bis(oxidanyl)oxan-2-yl]oxy-1-oxidanylidene-1-(pentylamino)propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propyl] ethanoate type: ligand / ID: 3 / Number of copies: 3 / Formula: JQ6 |
|---|---|
| Molecular weight | Theoretical: 840.975 Da |
| Chemical component information |  ChemComp-JQ6: |
-Macromolecule #4: [(2~{S})-3-[[(2~{S})-3-[(2~{S},3~{S},4~{S},5~{S},6~{S})-6-(hydrox...
| Macromolecule | Name: [(2~{S})-3-[[(2~{S})-3-[(2~{S},3~{S},4~{S},5~{S},6~{S})-6-(hydroxymethyl)-4,5-bis(oxidanyl)-3-(propanoylamino)oxan-2-yl]oxy-1-oxidanylidene-1-(pentadecylamino)propan-2-yl]oxy-oxidanyl- ...Name: [(2~{S})-3-[[(2~{S})-3-[(2~{S},3~{S},4~{S},5~{S},6~{S})-6-(hydroxymethyl)-4,5-bis(oxidanyl)-3-(propanoylamino)oxan-2-yl]oxy-1-oxidanylidene-1-(pentadecylamino)propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-2-octanoyloxy-propyl] decanoate type: ligand / ID: 4 / Number of copies: 3 / Formula: JPX |
|---|---|
| Molecular weight | Theoretical: 967.214 Da |
| Chemical component information | 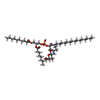 ChemComp-JPX: |
-Macromolecule #5: COPPER (II) ION
| Macromolecule | Name: COPPER (II) ION / type: ligand / ID: 5 / Number of copies: 9 / Formula: CU |
|---|---|
| Molecular weight | Theoretical: 63.546 Da |
| Chemical component information |  ChemComp-CU: |
-Macromolecule #6: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)