[English] 日本語
 Yorodumi
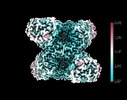
Yorodumi- EMDB-14587: Cryo-EM structure of the human GS-GN complex in the inhibited state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human GS-GN complex in the inhibited state | |||||||||
 Map data Map data | D2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycosyltransferase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) / glycogen(starch) synthase / glycogenin glucosyltransferase / Glycogen storage disease type II (GAA) / glycogenin glucosyltransferase activity / alpha-1,4-glucan glucosyltransferase (UDP-glucose donor) activity / D-glucose binding / glycogen biosynthetic process ...glycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) / glycogen(starch) synthase / glycogenin glucosyltransferase / Glycogen storage disease type II (GAA) / glycogenin glucosyltransferase activity / alpha-1,4-glucan glucosyltransferase (UDP-glucose donor) activity / D-glucose binding / glycogen biosynthetic process / Glycogen breakdown (glycogenolysis) / glycosyltransferase activity / inclusion body / Myoclonic epilepsy of Lafora / Glycogen synthesis / lysosomal lumen / manganese ion binding / heart development / secretory granule lumen / ficolin-1-rich granule lumen / Neutrophil degranulation / protein homodimerization activity / extracellular region / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
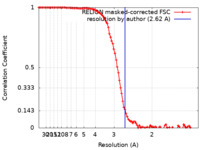
| Method | single particle reconstruction / cryo EM / Resolution: 2.62 Å | |||||||||
 Authors Authors | Marr L / Zeqiraj E | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Mechanism of glycogen synthase inactivation and interaction with glycogenin. Authors: Laura Marr / Dipsikha Biswas / Leonard A Daly / Christopher Browning / Sarah C M Vial / Daniel P Maskell / Catherine Hudson / Jay A Bertrand / John Pollard / Neil A Ranson / Heena Khatter / ...Authors: Laura Marr / Dipsikha Biswas / Leonard A Daly / Christopher Browning / Sarah C M Vial / Daniel P Maskell / Catherine Hudson / Jay A Bertrand / John Pollard / Neil A Ranson / Heena Khatter / Claire E Eyers / Kei Sakamoto / Elton Zeqiraj /   Abstract: Glycogen is the major glucose reserve in eukaryotes, and defects in glycogen metabolism and structure lead to disease. Glycogenesis involves interaction of glycogenin (GN) with glycogen synthase (GS) ...Glycogen is the major glucose reserve in eukaryotes, and defects in glycogen metabolism and structure lead to disease. Glycogenesis involves interaction of glycogenin (GN) with glycogen synthase (GS), where GS is activated by glucose-6-phosphate (G6P) and inactivated by phosphorylation. We describe the 2.6 Å resolution cryo-EM structure of phosphorylated human GS revealing an autoinhibited GS tetramer flanked by two GN dimers. Phosphorylated N- and C-termini from two GS protomers converge near the G6P-binding pocket and buttress against GS regulatory helices. This keeps GS in an inactive conformation mediated by phospho-Ser641 interactions with a composite "arginine cradle". Structure-guided mutagenesis perturbing interactions with phosphorylated tails led to increased basal/unstimulated GS activity. We propose that multivalent phosphorylation supports GS autoinhibition through interactions from a dynamic "spike" region, allowing a tuneable rheostat for regulating GS activity. This work therefore provides insights into glycogen synthesis regulation and facilitates studies of glycogen-related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14587.map.gz emd_14587.map.gz | 61.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14587-v30.xml emd-14587-v30.xml emd-14587.xml emd-14587.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14587_fsc.xml emd_14587_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14587.png emd_14587.png | 97.7 KB | ||
| Masks |  emd_14587_msk_1.map emd_14587_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14587.cif.gz emd-14587.cif.gz | 6.4 KB | ||
| Others |  emd_14587_additional_1.map.gz emd_14587_additional_1.map.gz emd_14587_half_map_1.map.gz emd_14587_half_map_1.map.gz emd_14587_half_map_2.map.gz emd_14587_half_map_2.map.gz | 61.1 MB 69.7 MB 69.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14587 http://ftp.pdbj.org/pub/emdb/structures/EMD-14587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14587 | HTTPS FTP |
-Related structure data
| Related structure data |  7zbnMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14587.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14587.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14587_msk_1.map emd_14587_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: C1
| File | emd_14587_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14587_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14587_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Glycogen synthase-Glycogenin complex
| Entire | Name: Glycogen synthase-Glycogenin complex |
|---|---|
| Components |
|
-Supramolecule #1: Glycogen synthase-Glycogenin complex
| Supramolecule | Name: Glycogen synthase-Glycogenin complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 485 KDa |
-Macromolecule #1: Glycogen [starch] synthase, muscle
| Macromolecule | Name: Glycogen [starch] synthase, muscle / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: glycogen(starch) synthase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.88543 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MPLNRTLSMS SLPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ ...String: MPLNRTLSMS SLPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ SEEKPHVVAH FHEWLAGVGL CLCRARRLPV ATIFTTHATL LGRYLCAGAV DFYNNLENFN VDKEAGERQI YH RYCMERA AAHCAHVFTT VSQITAIEAQ HLLKRKPDIV TPNGLNVKKF SAMHEFQNLH AQSKARIQEF VRGHFYGHLD FNL DKTLYF FIAGRYEFSN KGADVFLEAL ARLNYLLRVN GSEQTVVAFF IMPARTNNFN VETLKGQAVR KQLWDTANTV KEKF GRKLY ESLLVGSLPD MNKMLDKEDF TMMKRAIFAT QRQSFPPVCT HNMLDDSSDP ILTTIRRIGL FNSSADRVKV IFHPE FLSS TSPLLPVDYE EFVRGCHLGV FPSYYEPWGY TPAECTVMGI PSISTNLSGF GCFMEEHIAD PSAYGIYILD RRFRSL DDS CSQLTSFLYS FCQQSRRQRI IQRNRTERLS DLLDWKYLGR YYMSARHMAL SKAFPEHFTY EPNEADAAQG YRYPRPA SV PPSPSLSRHS SPHQSEDEED PRNGPLEEDG ERYDEDEEAA KDRRNIRAPE WPRRASCTSS TSGSKRNSVD TATSSSLS T PSEPLSPTSS LGEERN UniProtKB: Glycogen [starch] synthase, muscle |
-Macromolecule #2: Glycogen [starch] synthase, muscle
| Macromolecule | Name: Glycogen [starch] synthase, muscle / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: glycogen(starch) synthase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.965406 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MPLNRTLSMS SLPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ ...String: MPLNRTLSMS SLPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ SEEKPHVVAH FHEWLAGVGL CLCRARRLPV ATIFTTHATL LGRYLCAGAV DFYNNLENFN VDKEAGERQI YH RYCMERA AAHCAHVFTT VSQITAIEAQ HLLKRKPDIV TPNGLNVKKF SAMHEFQNLH AQSKARIQEF VRGHFYGHLD FNL DKTLYF FIAGRYEFSN KGADVFLEAL ARLNYLLRVN GSEQTVVAFF IMPARTNNFN VETLKGQAVR KQLWDTANTV KEKF GRKLY ESLLVGSLPD MNKMLDKEDF TMMKRAIFAT QRQSFPPVCT HNMLDDSSDP ILTTIRRIGL FNSSADRVKV IFHPE FLSS TSPLLPVDYE EFVRGCHLGV FPSYYEPWGY TPAECTVMGI PSISTNLSGF GCFMEEHIAD PSAYGIYILD RRFRSL DDS CSQLTSFLYS FCQQSRRQRI IQRNRTERLS DLLDWKYLGR YYMSARHMAL SKAFPEHFTY EPNEADAAQG YRYPRPA (SEP)V PPSPSLSRHS SPHQSEDEED PRNGPLEEDG ERYDEDEEAA KDRRNIRAPE WPRRASCTSS TSGSKRNSVD TATS SSLST PSEPLSPTSS LGEERN UniProtKB: Glycogen [starch] synthase, muscle |
-Macromolecule #3: Isoform GN-1 of Glycogenin-1
| Macromolecule | Name: Isoform GN-1 of Glycogenin-1 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO / EC number: glycogenin glucosyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.469348 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADQAFVTLT TNDAYAKGAL VLGSSLKQHR TTRRLVVLAT PQVSDSMRKV LETVFDEVIM VDVLDSGDSA HLTLMKRPEL GVTLTKLHC WSLTQYSKCV FMDADTLVLA NIDDLFDREE LSAAPDPGWP DCFNSGVFVY QPSVETYNQL LHLASEQGSF D GGDQGILN ...String: MADQAFVTLT TNDAYAKGAL VLGSSLKQHR TTRRLVVLAT PQVSDSMRKV LETVFDEVIM VDVLDSGDSA HLTLMKRPEL GVTLTKLHC WSLTQYSKCV FMDADTLVLA NIDDLFDREE LSAAPDPGWP DCFNSGVFVY QPSVETYNQL LHLASEQGSF D GGDQGILN TFFSSWATTD IRKHLPFIYN LSSISIFSYL PAFKVFGASA KVVHFLGRVK PWNYTYDPKT KSVKSEAHDP NM THPEFLI LWWNIFTTNV LPLLQQFGLV KDTCSYVNVE DVSGAISHLS LGEIPAMAQP FVSSEERKER WEQGQADYMG ADS FDNIKR KLDTYLQ UniProtKB: Glycogenin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 34.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)