+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tick-borne encephalitis virus Kuutsalo-14 | ||||||||||||||||||||||||||||||
 Map data Map data | Automatically locally sharpened map used for model buidling. Sharpened using Xmipp Localdeblursharpening. | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | virion / membrane protein / glycoprotein / VIRUS | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationflavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral capsid / nucleoside-triphosphate phosphatase / double-stranded RNA binding / clathrin-dependent endocytosis of virus by host cell / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / methyltransferase cap1 activity ...flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral capsid / nucleoside-triphosphate phosphatase / double-stranded RNA binding / clathrin-dependent endocytosis of virus by host cell / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / symbiont-mediated suppression of host innate immune response / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / serine-type endopeptidase activity / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / metal ion binding / membrane Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  Tick-borne encephalitis virus (WESTERN SUBTYPE) / Tick-borne encephalitis virus (WESTERN SUBTYPE) /  Tick-borne encephalitis virus Tick-borne encephalitis virus | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Pulkkinen LIA / Barrass SV / Anastasina M / Butcher SJ | ||||||||||||||||||||||||||||||
| Funding support |  Finland, European Union, Finland, European Union,  Sweden, 9 items Sweden, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Molecular Organisation of Tick-Borne Encephalitis Virus. Authors: Lauri I A Pulkkinen / Sarah V Barrass / Aušra Domanska / Anna K Överby / Maria Anastasina / Sarah J Butcher /   Abstract: Tick-borne encephalitis virus (TBEV) is a pathogenic, enveloped, positive-stranded RNA virus in the family . Structural studies of flavivirus virions have primarily focused on mosquito-borne species, ...Tick-borne encephalitis virus (TBEV) is a pathogenic, enveloped, positive-stranded RNA virus in the family . Structural studies of flavivirus virions have primarily focused on mosquito-borne species, with only one cryo-electron microscopy (cryo-EM) structure of a tick-borne species published. Here, we present a 3.3 Å cryo-EM structure of the TBEV virion of the Kuutsalo-14 isolate, confirming the overall organisation of the virus. We observe conformational switching of the peripheral and transmembrane helices of M protein, which can explain the quasi-equivalent packing of the viral proteins and highlights their importance in stabilising membrane protein arrangement in the virion. The residues responsible for M protein interactions are highly conserved in TBEV but not in the structurally studied Hypr strain, nor in mosquito-borne flaviviruses. These interactions may compensate for the lower number of hydrogen bonds between E proteins in TBEV compared to the mosquito-borne flaviviruses. The structure reveals two lipids bound in the E protein which are important for virus assembly. The lipid pockets are comparable to those recently described in mosquito-borne Zika, Spondweni, Dengue, and Usutu viruses. Our results thus advance the understanding of tick-borne flavivirus architecture and virion-stabilising interactions. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14512.map.gz emd_14512.map.gz | 785 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14512-v30.xml emd-14512-v30.xml emd-14512.xml emd-14512.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14512_fsc.xml emd_14512_fsc.xml | 22.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14512.png emd_14512.png | 213.6 KB | ||
| Filedesc metadata |  emd-14512.cif.gz emd-14512.cif.gz | 6.2 KB | ||
| Others |  emd_14512_additional_1.map.gz emd_14512_additional_1.map.gz emd_14512_half_map_1.map.gz emd_14512_half_map_1.map.gz emd_14512_half_map_2.map.gz emd_14512_half_map_2.map.gz | 522.4 MB 810.1 MB 808.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14512 http://ftp.pdbj.org/pub/emdb/structures/EMD-14512 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14512 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14512 | HTTPS FTP |
-Related structure data
| Related structure data |  7z51MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11014 (Title: Single-particle reconstruction of Tick-borne encephalitis virus (Final reconstruction) EMPIAR-11014 (Title: Single-particle reconstruction of Tick-borne encephalitis virus (Final reconstruction)Data size: 16.4 TB Data #1: Preparation 2 unaligned movies [micrographs - multiframe] Data #2: Preparation 3 unaligned movies [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14512.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14512.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Automatically locally sharpened map used for model buidling. Sharpened using Xmipp Localdeblursharpening. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
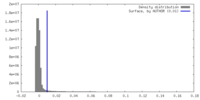
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Minimally sharpened map used for model building. B factor -5 A^2.
| File | emd_14512_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Minimally sharpened map used for model building. B factor -5 A^2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
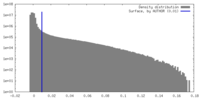
| Density Histograms |
-Half map: #1
| File | emd_14512_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
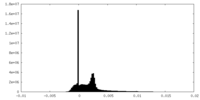
| Density Histograms |
-Half map: #2
| File | emd_14512_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tick-borne encephalitis virus
| Entire | Name:  Tick-borne encephalitis virus Tick-borne encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Tick-borne encephalitis virus
| Supramolecule | Name: Tick-borne encephalitis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Virus was produced in SK-N-SH cells, purified, and inactivated with UV-C. NCBI-ID: 11088 / Sci species name: Tick-borne encephalitis virus / Sci species strain: Neudoerfl / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope protein E
| Macromolecule | Name: Envelope protein E / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tick-borne encephalitis virus (WESTERN SUBTYPE) / Strain: Neudoerfl Tick-borne encephalitis virus (WESTERN SUBTYPE) / Strain: Neudoerfl |
| Molecular weight | Theoretical: 53.55827 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SRCTHLENRD FVTGTQGTTR VTLVLELGGC VTITAEGKPS MDVWLDAIYQ ENPAKTREYC LHAKLSDTKV AARCPTMGPA TLAEEHQGG TVCKRDQSDR GWGNHCGLFG KGSIVACVKA ACEAKKKATG HVYDANKIVY TVKVEPHTGD YVAANETHSG R KTASFTVS ...String: SRCTHLENRD FVTGTQGTTR VTLVLELGGC VTITAEGKPS MDVWLDAIYQ ENPAKTREYC LHAKLSDTKV AARCPTMGPA TLAEEHQGG TVCKRDQSDR GWGNHCGLFG KGSIVACVKA ACEAKKKATG HVYDANKIVY TVKVEPHTGD YVAANETHSG R KTASFTVS SEKTILTMGE YGDVSLLCRV ASGVDLAQTV ILELDKTVEH LPTAWQVHRD WFNDLALPWK HEGAQNWNNA ER LVEFGAP HAVKMDVYNL GDQTGVLLKA LAGVPVAHIE GTKYHLKSGH VTCEVGLEKL KMKGLTYTMC DKTKFTWKRA PTD SGHDTV VMEVTFSGTK PCRIPVRAVA HGSPDVNVAM LITPNPTIEN NGGGFIEMQL PPGDNIIYVG ELSHQWFQKG SSIG RVFQK TKKGIERLTV IGEHAWDFGS AGGFLSSIGK AVHTVLGGAF NSIFGGVGFL PKLLLGVALA WLGLNMRNPT MSMSF LLAG GLVLAMTLGV GA UniProtKB: Genome polyprotein |
-Macromolecule #2: Small envelope protein M
| Macromolecule | Name: Small envelope protein M / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tick-borne encephalitis virus (WESTERN SUBTYPE) / Strain: Neudoerfl Tick-borne encephalitis virus (WESTERN SUBTYPE) / Strain: Neudoerfl |
| Molecular weight | Theoretical: 8.311854 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SVLIPSHAQG ELTGRGHKWL EGDSLRTHLT RVEGWVWKNK LLALAMVTVV WLTLESVVTR VAVLVVLLCL APVYA UniProtKB: Genome polyprotein |
-Macromolecule #4: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 4 / Number of copies: 6 / Formula: CPL |
|---|---|
| Molecular weight | Theoretical: 758.06 Da |
| Chemical component information |  ChemComp-CPL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)