[English] 日本語
 Yorodumi
Yorodumi- EMDB-14468: Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A | |||||||||||||||||||||
 Map data Map data | Cryo-EM structure of the Ty1 integrase-RNA Polymerase III post cleavage complex (Pol III PCC) at 3.1 A | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | targeted DNA integration / Ty1 retrotransposon / Ty1 integrase / RNA 19 polymerase III / transcription | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtransposition / RNA Polymerase I Transcription Initiation / ribonuclease H / Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes ...transposition / RNA Polymerase I Transcription Initiation / ribonuclease H / Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / termination of RNA polymerase III transcription / RNA Polymerase II Pre-transcription Events / RNA-templated transcription / Formation of TC-NER Pre-Incision Complex / RNA Polymerase I Promoter Escape / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / Gap-filling DNA repair synthesis and ligation in TC-NER / nucleolar large rRNA transcription by RNA polymerase I / transcription initiation at RNA polymerase I promoter / Estrogen-dependent gene expression / transcription by RNA polymerase III / Dual incision in TC-NER / RNA polymerase I complex / RNA polymerase III complex / transcription elongation by RNA polymerase I / tRNA transcription by RNA polymerase III / RNA polymerase II, core complex / transcription by RNA polymerase I / nucleotidyltransferase activity / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / DNA integration / RNA-directed DNA polymerase / ribonucleoside binding / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / peroxisome / single-stranded DNA binding / ribosome biogenesis / DNA recombination / DNA-directed DNA polymerase / nucleic acid binding / aspartic-type endopeptidase activity / transcription by RNA polymerase II / DNA-directed DNA polymerase activity / protein dimerization activity / viral translational frameshifting / nucleotide binding / nucleolus / mitochondrion / proteolysis / DNA binding / RNA binding / zinc ion binding / nucleoplasm / ATP binding / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
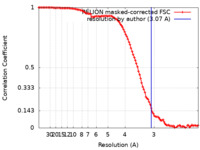
| Method | single particle reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||||||||||||||
 Authors Authors | Nguyen PQ / Fernandez-Tornero C | |||||||||||||||||||||
| Funding support | European Union,  France, France,  Spain, 6 items Spain, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration. Authors: Phong Quoc Nguyen / Sonia Huecas / Amna Asif-Laidin / Adrián Plaza-Pegueroles / Beatrice Capuzzi / Noé Palmic / Christine Conesa / Joël Acker / Juan Reguera / Pascale Lesage / Carlos Fernández-Tornero /   Abstract: The yeast Ty1 retrotransposon integrates upstream of genes transcribed by RNA polymerase III (Pol III). Specificity of integration is mediated by an interaction between the Ty1 integrase (IN1) and ...The yeast Ty1 retrotransposon integrates upstream of genes transcribed by RNA polymerase III (Pol III). Specificity of integration is mediated by an interaction between the Ty1 integrase (IN1) and Pol III, currently uncharacterized at the atomic level. We report cryo-EM structures of Pol III in complex with IN1, revealing a 16-residue segment at the IN1 C-terminus that contacts Pol III subunits AC40 and AC19, an interaction that we validate by in vivo mutational analysis. Binding to IN1 associates with allosteric changes in Pol III that may affect its transcriptional activity. The C-terminal domain of subunit C11, involved in RNA cleavage, inserts into the Pol III funnel pore, providing evidence for a two-metal mechanism during RNA cleavage. Additionally, ordering next to C11 of an N-terminal portion from subunit C53 may explain the connection between these subunits during termination and reinitiation. Deletion of the C53 N-terminal region leads to reduced chromatin association of Pol III and IN1, and a major fall in Ty1 integration events. Our data support a model in which IN1 binding induces a Pol III configuration that may favor its retention on chromatin, thereby improving the likelihood of Ty1 integration. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14468.map.gz emd_14468.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14468-v30.xml emd-14468-v30.xml emd-14468.xml emd-14468.xml | 52.3 KB 52.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14468_fsc.xml emd_14468_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14468.png emd_14468.png | 20.4 KB | ||
| Filedesc metadata |  emd-14468.cif.gz emd-14468.cif.gz | 12.4 KB | ||
| Others |  emd_14468_half_map_1.map.gz emd_14468_half_map_1.map.gz emd_14468_half_map_2.map.gz emd_14468_half_map_2.map.gz | 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14468 http://ftp.pdbj.org/pub/emdb/structures/EMD-14468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14468 | HTTPS FTP |
-Related structure data
| Related structure data |  7z2zMC  7z0hC  7z30C  7z31C  8bwsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14468.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14468.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the Ty1 integrase-RNA Polymerase III post cleavage complex (Pol III PCC) at 3.1 A | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.085 Å | ||||||||||||||||||||||||||||||||||||
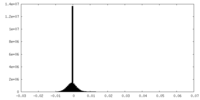
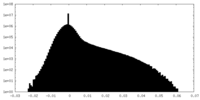
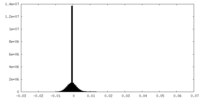
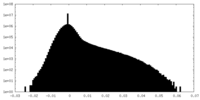
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM half-map2 of the Ty1 integrase-RNA Polymerase III...
| File | emd_14468_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half-map2 of the Ty1 integrase-RNA Polymerase III post cleavage complex (Pol III PCC) at 3.1 A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM half-map1 of the Ty1 integrase-RNA Polymerase III...
| File | emd_14468_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half-map1 of the Ty1 integrase-RNA Polymerase III post cleavage complex (Pol III PCC) at 3.1 A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of yeast RNA Polymerase III - DNA - Ty1 integrase (Pol ...
+Supramolecule #1: Structure of yeast RNA Polymerase III - DNA - Ty1 integrase (Pol ...
+Supramolecule #2: RNA Polymerase III - Ty1 integrase
+Supramolecule #3: RNA, DNA
+Macromolecule #1: DNA-directed RNA polymerase III subunit RPC1
+Macromolecule #2: DNA-directed RNA polymerase III subunit RPC2
+Macromolecule #3: DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #4: DNA-directed RNA polymerase III subunit RPC9
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerase III subunit RPC8
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase III subunit RPC10
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #11: DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #13: DNA-directed RNA polymerase III subunit RPC5
+Macromolecule #14: DNA-directed RNA polymerase III subunit RPC4
+Macromolecule #15: DNA-directed RNA polymerase III subunit RPC3
+Macromolecule #16: DNA-directed RNA polymerase III subunit RPC6
+Macromolecule #17: DNA-directed RNA polymerase III subunit RPC7
+Macromolecule #21: Integrase
+Macromolecule #22: Unknown RNA polymerase III chain
+Macromolecule #18: RNA
+Macromolecule #19: Non-template DNA
+Macromolecule #20: Template DNA
+Macromolecule #23: ZINC ION
+Macromolecule #24: MAGNESIUM ION
+Macromolecule #25: (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pent...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 42.45 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)