[English] 日本語
 Yorodumi
Yorodumi- EMDB-14461: P. falciparum kinesin-8B motor domain in no nucleotide bound to t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 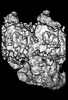 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
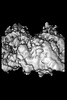
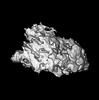
| Title | P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinesin / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMicrotubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / microtubule motor activity / Recruitment of NuMA to mitotic centrosomes / COPI-mediated anterograde transport / microtubule-based movement / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / microtubule cytoskeleton / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / GTPase activity / GTP binding / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |    | |||||||||
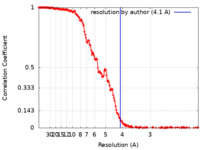
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Liu T / Shilliday F / Cook AD / Moores CA | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation | Journal: Protein Sci / Year: 2018 Title: UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Authors: Thomas D Goddard / Conrad C Huang / Elaine C Meng / Eric F Pettersen / Gregory S Couch / John H Morris / Thomas E Ferrin /  Abstract: UCSF ChimeraX is next-generation software for the visualization and analysis of molecular structures, density maps, 3D microscopy, and associated data. It addresses challenges in the size, scope, and ...UCSF ChimeraX is next-generation software for the visualization and analysis of molecular structures, density maps, 3D microscopy, and associated data. It addresses challenges in the size, scope, and disparate types of data attendant with cutting-edge experimental methods, while providing advanced options for high-quality rendering (interactive ambient occlusion, reliable molecular surface calculations, etc.) and professional approaches to software design and distribution. This article highlights some specific advances in the areas of visualization and usability, performance, and extensibility. ChimeraX is free for noncommercial use and is available from http://www.rbvi.ucsf.edu/chimerax/ for Windows, Mac, and Linux. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14461.map.gz emd_14461.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14461-v30.xml emd-14461-v30.xml emd-14461.xml emd-14461.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14461_fsc.xml emd_14461_fsc.xml | 19.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14461.png emd_14461.png | 84 KB | ||
| Filedesc metadata |  emd-14461.cif.gz emd-14461.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14461 http://ftp.pdbj.org/pub/emdb/structures/EMD-14461 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14461 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14461 | HTTPS FTP |
-Validation report
| Summary document |  emd_14461_validation.pdf.gz emd_14461_validation.pdf.gz | 414.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14461_full_validation.pdf.gz emd_14461_full_validation.pdf.gz | 413.9 KB | Display | |
| Data in XML |  emd_14461_validation.xml.gz emd_14461_validation.xml.gz | 11.4 KB | Display | |
| Data in CIF |  emd_14461_validation.cif.gz emd_14461_validation.cif.gz | 16.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14461 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14461 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14461 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14461 | HTTPS FTP |
-Related structure data
| Related structure data |  7z2cMC  7z2aC  7z2bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14461.map.gz / Format: CCP4 / Size: 4.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14461.map.gz / Format: CCP4 / Size: 4.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
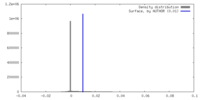
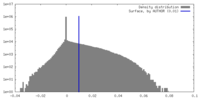
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : P. falciparum kinesin-8B motor domain in no nucleotide state boun...
| Entire | Name: P. falciparum kinesin-8B motor domain in no nucleotide state bound to tubulin dimer |
|---|---|
| Components |
|
-Supramolecule #1: P. falciparum kinesin-8B motor domain in no nucleotide state boun...
| Supramolecule | Name: P. falciparum kinesin-8B motor domain in no nucleotide state bound to tubulin dimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Kinesin-like protein
| Macromolecule | Name: Kinesin-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.137656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DITYNMNVVI RCRPMSISEK NDGAKNVIKI LENKMVVLLD PSDNSDNVLR QNRSREKKYV FDYVFDENSS QEDVYNNSVK CLIDAVIKG YNSTVFAYGA TGAGKTHTII GYKNEPGIMM MILKDLFERI KMLQIMNEYK VKCSFIEIYN ENICDLLNPS N EYLDVRED ...String: DITYNMNVVI RCRPMSISEK NDGAKNVIKI LENKMVVLLD PSDNSDNVLR QNRSREKKYV FDYVFDENSS QEDVYNNSVK CLIDAVIKG YNSTVFAYGA TGAGKTHTII GYKNEPGIMM MILKDLFERI KMLQIMNEYK VKCSFIEIYN ENICDLLNPS N EYLDVRED PIKGVTVSNI FEVCTTSVEE IMELIHTGNR NRTQEPTDAN KTSSRSHGVL QVIVEETEKG QGVYQQTKRG KL CVIDLAG SERASQTNNK GMRMLEGANI NRSLLALGNV INALVLRSKG NSKSNFIPFR DSKLTRLLKD SLGGNCKTVM IAN ISPSHL SYEDTHNTLK YANRAKNIKN V UniProtKB: Kinesin-like protein |
-Macromolecule #2: Detyrosinated tubulin alpha-1B chain
| Macromolecule | Name: Detyrosinated tubulin alpha-1B chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.833184 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPDSF NTFFSETGAG KHVPRAVFVD LEPTVIDEVR TGTYRQLFHP EQLITGKED AANNYARGHY TIGKEIIDLV LDRIRKLADQ CTGLQGFLVF HSFGGGTGSG FTSLLMERLS VDYGKKSKLE F SIYPAPQV ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPDSF NTFFSETGAG KHVPRAVFVD LEPTVIDEVR TGTYRQLFHP EQLITGKED AANNYARGHY TIGKEIIDLV LDRIRKLADQ CTGLQGFLVF HSFGGGTGSG FTSLLMERLS VDYGKKSKLE F SIYPAPQV STAVVEPYNS ILTTHTTLEH SDCAFMVDNE AIYDICRRNL DIERPTYTNL NRLISQIVSS ITASLRFDGA LN VDLTEFQ TNLVPYPRIH FPLATYAPVI SAEKAYHEQL SVAEITNACF EPANQMVKCD PRHGKYMACC LLYRGDVVPK DVN AAIATI KTKRSIQFVD WCPTGFKVGI NYQPPTVVPG GDLAKVQRAV CMLSNTTAIA EAWARLDHKF DLMYAKRAFV HWYV GEGME EGEFSEARED MAALEKDYEE VGV UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #3: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.825859 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY Q UniProtKB: Tubulin beta chain |
-Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 6 / Number of copies: 1 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 46.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


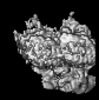







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)