+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pol III PTC - Full transcription bubble | |||||||||
 Map data Map data | Pol III PTC NTPs DNA/RNA classified Sharpened with LocalDeblur | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA synthesis / short RNAs / termination / TRANSCRIPTION | |||||||||
| Biological species |  | |||||||||
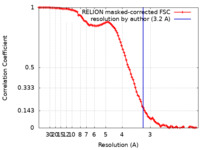
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Girbig M / Mueller CW | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals. Authors: Mathias Girbig / Juanjuan Xie / Helga Grötsch / Domenico Libri / Odil Porrua / Christoph W Müller /   Abstract: RNA polymerase (Pol) III is specialized to transcribe short, abundant RNAs, for which it terminates transcription on polythymine (dT) stretches on the non-template (NT) strand. When Pol III reaches ...RNA polymerase (Pol) III is specialized to transcribe short, abundant RNAs, for which it terminates transcription on polythymine (dT) stretches on the non-template (NT) strand. When Pol III reaches the termination signal, it pauses and forms the pre-termination complex (PTC). Here, we report cryoelectron microscopy (cryo-EM) structures of the yeast Pol III PTC and complementary functional states at resolutions of 2.7-3.9 Å. Pol III recognizes the poly(dT) termination signal with subunit C128 that forms a hydrogen-bond network with the NT strand and, thereby, induces pausing. Mutating key interacting residues interferes with transcription termination in vitro, impairs yeast growth, and causes global termination defects in vivo, confirming our structural results. Additional cryo-EM analysis reveals that C53-C37, a Pol III subcomplex and key termination factor, participates indirectly in Pol III termination. We propose a mechanistic model of Pol III transcription termination and rationalize why Pol III, unlike Pol I and Pol II, terminates on poly(dT) signals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14450.map.gz emd_14450.map.gz | 152.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14450-v30.xml emd-14450-v30.xml emd-14450.xml emd-14450.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14450_fsc.xml emd_14450_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_14450.png emd_14450.png | 69.1 KB | ||
| Filedesc metadata |  emd-14450.cif.gz emd-14450.cif.gz | 4.4 KB | ||
| Others |  emd_14450_additional_1.map.gz emd_14450_additional_1.map.gz | 152.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14450 http://ftp.pdbj.org/pub/emdb/structures/EMD-14450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14450 | HTTPS FTP |
-Validation report
| Summary document |  emd_14450_validation.pdf.gz emd_14450_validation.pdf.gz | 460.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14450_full_validation.pdf.gz emd_14450_full_validation.pdf.gz | 459.9 KB | Display | |
| Data in XML |  emd_14450_validation.xml.gz emd_14450_validation.xml.gz | 11.9 KB | Display | |
| Data in CIF |  emd_14450_validation.cif.gz emd_14450_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14450 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14450 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14450.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14450.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pol III PTC NTPs DNA/RNA classified Sharpened with LocalDeblur | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.041 Å | ||||||||||||||||||||||||||||||||||||
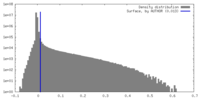
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Pol III PTC NTPs DNA/RNA classified Unsharpened map
| File | emd_14450_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pol III PTC NTPs DNA/RNA classified Unsharpened map | ||||||||||||
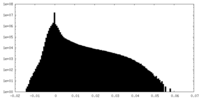
| Projections & Slices |
| ||||||||||||
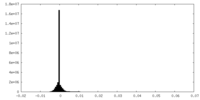
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast RNA Polymerase III PTC + NTPs, Full transcription bubble
| Entire | Name: Yeast RNA Polymerase III PTC + NTPs, Full transcription bubble |
|---|---|
| Components |
|
-Supramolecule #1: Yeast RNA Polymerase III PTC + NTPs, Full transcription bubble
| Supramolecule | Name: Yeast RNA Polymerase III PTC + NTPs, Full transcription bubble type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#20 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 730 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.4 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 IS (4k x 4k) / Average electron dose: 44.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Protocol: OTHER |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)