+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
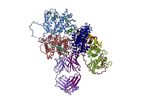
| Title | VAR2 complex with PAM1.4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VAR2CSA / malaria / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
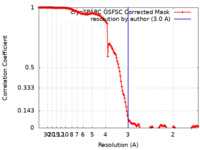
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Raghavan SSR / Wang KT | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2022 Journal: PLoS Pathog / Year: 2022Title: Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA. Authors: Sai Sundar Rajan Raghavan / Robert Dagil / Mary Lopez-Perez / Julian Conrad / Maria Rosaria Bassi / Maria Del Pilar Quintana / Swati Choudhary / Tobias Gustavsson / Yong Wang / Pontus ...Authors: Sai Sundar Rajan Raghavan / Robert Dagil / Mary Lopez-Perez / Julian Conrad / Maria Rosaria Bassi / Maria Del Pilar Quintana / Swati Choudhary / Tobias Gustavsson / Yong Wang / Pontus Gourdon / Michael Fokuo Ofori / Sebastian Boje Christensen / Daniel Thomas Remias Minja / Christentze Schmiegelow / Morten Agertoug Nielsen / Lea Barfod / Lars Hviid / Ali Salanti / Thomas Lavstsen / Kaituo Wang /    Abstract: Malaria during pregnancy is a major global health problem caused by infection with Plasmodium falciparum parasites. Severe effects arise from the accumulation of infected erythrocytes in the placenta. ...Malaria during pregnancy is a major global health problem caused by infection with Plasmodium falciparum parasites. Severe effects arise from the accumulation of infected erythrocytes in the placenta. Here, erythrocytes infected by late blood-stage parasites adhere to placental chondroitin sulphate A (CS) via VAR2CSA-type P. falciparum erythrocyte membrane protein 1 (PfEMP1) adhesion proteins. Immunity to placental malaria is acquired through exposure and mediated through antibodies to VAR2CSA. Through evolution, the VAR2CSA proteins have diversified in sequence to escape immune recognition but retained their overall macromolecular structure to maintain CS binding affinity. This structural conservation may also have allowed development of broadly reactive antibodies to VAR2CSA in immune women. Here we show the negative stain and cryo-EM structure of the only known broadly reactive human monoclonal antibody, PAM1.4, in complex with VAR2CSA. The data shows how PAM1.4's broad VAR2CSA reactivity is achieved through interactions with multiple conserved residues of different sub-domains forming conformational epitope distant from the CS binding site on the VAR2CSA core structure. Thus, while PAM1.4 may represent a class of antibodies mediating placental malaria immunity by inducing phagocytosis or NK cell-mediated cytotoxicity, it is likely that broadly CS binding-inhibitory antibodies target other epitopes at the CS binding site. Insights on both types of broadly reactive monoclonal antibodies may aid the development of a vaccine against placental malaria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14438.map.gz emd_14438.map.gz | 166.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14438-v30.xml emd-14438-v30.xml emd-14438.xml emd-14438.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14438_fsc.xml emd_14438_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14438.png emd_14438.png | 64.7 KB | ||
| Filedesc metadata |  emd-14438.cif.gz emd-14438.cif.gz | 7.7 KB | ||
| Others |  emd_14438_additional_1.map.gz emd_14438_additional_1.map.gz | 307.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14438 http://ftp.pdbj.org/pub/emdb/structures/EMD-14438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14438 | HTTPS FTP |
-Related structure data
| Related structure data |  7z12MC  7z1hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14438.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14438.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_14438_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VAR2CSA complex with PAM1.4
| Entire | Name: VAR2CSA complex with PAM1.4 |
|---|---|
| Components |
|
-Supramolecule #1: VAR2CSA complex with PAM1.4
| Supramolecule | Name: VAR2CSA complex with PAM1.4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: PAM1.4, Heavy Chain
| Macromolecule | Name: PAM1.4, Heavy Chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.554219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHSE VRLVEYGGRV VRPGGSLRLS CAAGGFDFDD YGMSWVRQAP GKGLEWVAGI NWNALDKKYA DSVKGRFTI SRDNPKSSVY LQMTSLTAED TALYYCARDL RNSIFATGAL GNWGQGTLVI VSSASTKGPS VFPLAPSSKS T SGGTAALG ...String: MGWSCIILFL VATATGVHSE VRLVEYGGRV VRPGGSLRLS CAAGGFDFDD YGMSWVRQAP GKGLEWVAGI NWNALDKKYA DSVKGRFTI SRDNPKSSVY LQMTSLTAED TALYYCARDL RNSIFATGAL GNWGQGTLVI VSSASTKGPS VFPLAPSSKS T SGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KV EPKSCDK THTCPPCPAP ELLGGPSVFL FPPKPKDTLM ISRTPEVTCV VVDVSHEDPE VKFNWYVDGV EVHNAKTKPR EEQ YNSTYR VVSVLTVLHQ DWLNGKEYKC KVSNKALPAP IEKTISKAKG QPREPQVYTL PPSRDELTKN QVSLTCLVKG FYPS DIAVE WESNGQPENN YKTTPPVLDS DGSFFLYSKL TVDKSRWQQG NVFSCSVMHE ALHNHYTQKS LSLSPGK |
-Macromolecule #2: PAM1.4, light Chain
| Macromolecule | Name: PAM1.4, light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.425562 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHCD IQMTQSPSSL SASIGDRVTI TCRASQDIAN YLAWYQQKPG TVPKLLIYAA STLLSGVPSR FSGRQSGTH FTLTISSLQP EDVATYYCQK YNNAPAAFGQ GTRLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY P REAKVQWK ...String: MGWSCIILFL VATATGVHCD IQMTQSPSSL SASIGDRVTI TCRASQDIAN YLAWYQQKPG TVPKLLIYAA STLLSGVPSR FSGRQSGTH FTLTISSLQP EDVATYYCQK YNNAPAAFGQ GTRLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY P REAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Macromolecule #3: VAR2CSA
| Macromolecule | Name: VAR2CSA / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: FVO |
| Molecular weight | Theoretical: 235.359453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSTSTIANK IEEYLGAKSD DSKIDELLKA DPSEVEYYRS GGDGDYLKNN ICKITVNHSD SGKYDPCEKK LPPYDDNDQW KCQQNSSDG SGKPENICVP PRRERLCTYN LENLKFDKIR DNNAFLADVL LTARNEGEKI VQNHPDTNSS NVCNALERSF A DLADIIRG ...String: MDSTSTIANK IEEYLGAKSD DSKIDELLKA DPSEVEYYRS GGDGDYLKNN ICKITVNHSD SGKYDPCEKK LPPYDDNDQW KCQQNSSDG SGKPENICVP PRRERLCTYN LENLKFDKIR DNNAFLADVL LTARNEGEKI VQNHPDTNSS NVCNALERSF A DLADIIRG TDQWKGTNSN LEKNLKQMFA KIRENDKVLQ DKYPKDQKYT KLREAWWNAN RQKVWEVITC GARSNDLLIK RG WRTSGKS DRKKNFELCR KCGHYEKEVP TKLDYVPQFL RWLTEWIEDF YREKQNLIDD MERHREECTR EDHKSKEGTS YCS TCKDKC KKYCECVKKW KTEWENQENK YKDLYEQNKN KTSQKNTSRY DDYVKDFFEK LEANYSSLEN YIKGDPYFAE YATK LSFIL NPSDANNPSG ETANHNDEAC NCNESGISSV GQAQTSGPSS NKTCITHSSI KTNKKKECKD VKLGVRENDK DLKIC VIED TSLSGVDNCC CQDLLGILQE NCSDNKRGSS SNDSCDNKNQ DECQKKLEKV FASLTNGYKC DKCKSGTSRS KKKWIW KKS SGNEEGLQEE YANTIGLPPR TQSLYLGNLP KLENVCEDVK DINFDTKEKF LAGCLIVSFH EGKNLKKRYP QNKNSGN KE NLCKALEYSF ADYGDLIKGT SIWDNEYTKD LELNLQNNFG KLFGKYIKKN NTAEQDTSYS SLDELRESWW NTNKKYIW T AMKHGAEMNI TTCNADGSVT GSGSSCDDIP TIDLIPQYLR FLQEWVENFC EQRQAKVKDV ITNCKSCKES GNKCKTECK TKCKDECEKY KKFIEACGTA GGGIGTAGSP WSKRWDQIYK RYSKHIEDAK RNRKAGTKNC GTSSTTNAAA STDENKCVQS DIDSFFKHL IDIGLTTPSS YLSNVLDDNI CGADKAPWTT YTTYTTTEKC NKERDKSKSQ SSDTLVVVNV PSPLGNTPYR Y KYACQCKI PTNEETCDDR KEYMNQWSCG SARTMKRGYK NDNYELCKYN GVDVKPTTVR SNSSKLDGND VTFFNLFEQW NK EIQYQIE QYMTNANISC IDEKEVLDSV SDEGTPKVRG GYEDGRNNNT DQGTNCKEKC KCYKLWIEKI NDQWGKQKDN YNK FRSKQI YDANKGSQNK KVVSLSNFLF FSCWEEYIQK YFNGDWSKIK NIGSDTFEFL IKKCGNNSAH GEEIFSEKLK NAEK KCKEN ESTDTNINKS ETSCDLNATN YIRGCQSKTY DGKIFPGKGG EKQWICKDTI IHGDTNGACI PPRTQNLCVG ELWDK SYGG RSNIKNDTKE LLKEKIKNAI HKETELLYEY HDTGTAIISK NDKKGQKGKN DPNGLPKGFC HAVQRSFIDY KNMILG TSV NIYEHIGKLQ EDIKKIIEKG TPQQKDKIGG VGSSTENVNA WWKGIEREMW DAVRCAITKI NKKNNNSIFN GDECGVS PP TGNDEDQSVS WFKEWGEQFC IERLRYEQNI REACTINGKN EKKCINSKSG QGDKIQGACK RKCEKYKKYI SEKKQEWD K QKTKYENKYV GKSASDLLKE NYPECISANF DFIFNDNIEY KTYYPYGDYS SICSCEQVKY YKYNNAEKKN NKSLCYEKD NDMTWSKKYI KKLENGRSLE GVYVPPRRQQ LCLYELFPII IKNEEGMEKA KEELLETLQI VAEREAYYLW KQYNPTGKGI DDANKKACC AIRGSFYDLE DIIKGNDLVH DEYTKYIDSK LNEIFGSSNT NDIDTKRART DWWENETITN GTDRKTIRQL V WDAMQSGV RYAVEEKNEN FPLCMGVEHI GIAKPQFIRW LEEWTNEFCE KYTKYFEDMK SKCDPPKRAD TCGDNSNIEC KK ACANYTN WLNPKRIEWN GMSNYYNKIY RKSNKESEDG KDYSMIMAPT VIDYLNKRCH GEINGNYICC SCKNIGAYNT TSG TVNKKL QKKETECEEE KGPLDLMNEV LNKMDKKYSA HKMKCTEVYL EHVEEQLNEI DNAIKDYKLY PLDRCFDDQT KMKV CDLIA DAIGCKDKTK LDELDEWNDM DLRGTYNKHK GVLIP UniProtKB: Erythrocyte membrane protein 1, PfEMP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)