+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14232 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Beta Variant SARS-CoV-2 Spike with 1 Erect RBD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spike / SARS-CoV-2 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Benton DJ / Wrobel AG | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Evolution of the SARS-CoV-2 spike protein in the human host. Authors: Antoni G Wrobel / Donald J Benton / Chloë Roustan / Annabel Borg / Saira Hussain / Stephen R Martin / Peter B Rosenthal / John J Skehel / Steven J Gamblin /  Abstract: Recently emerged variants of SARS-CoV-2 contain in their surface spike glycoproteins multiple substitutions associated with increased transmission and resistance to neutralising antibodies. We have ...Recently emerged variants of SARS-CoV-2 contain in their surface spike glycoproteins multiple substitutions associated with increased transmission and resistance to neutralising antibodies. We have examined the structure and receptor binding properties of spike proteins from the B.1.1.7 (Alpha) and B.1.351 (Beta) variants to better understand the evolution of the virus in humans. Spikes of both variants have the same mutation, N501Y, in the receptor-binding domains. This substitution confers tighter ACE2 binding, dependent on the common earlier substitution, D614G. Each variant spike has acquired other key changes in structure that likely impact virus pathogenesis. The spike from the Alpha variant is more stable against disruption upon binding ACE2 receptor than all other spikes studied. This feature is linked to the acquisition of a more basic substitution at the S1-S2 furin site (also observed for the variants of concern Delta, Kappa, and Omicron) which allows for near-complete cleavage. In the Beta variant spike, the presence of a new substitution, K417N (also observed in the Omicron variant), in combination with the D614G, stabilises a more open spike trimer, a conformation required for receptor binding. Our observations suggest ways these viruses have evolved to achieve greater transmissibility in humans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14232.map.gz emd_14232.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14232-v30.xml emd-14232-v30.xml emd-14232.xml emd-14232.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14232_fsc.xml emd_14232_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14232.png emd_14232.png | 113.5 KB | ||
| Masks |  emd_14232_msk_1.map emd_14232_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14232.cif.gz emd-14232.cif.gz | 6.9 KB | ||
| Others |  emd_14232_half_map_1.map.gz emd_14232_half_map_1.map.gz emd_14232_half_map_2.map.gz emd_14232_half_map_2.map.gz | 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14232 http://ftp.pdbj.org/pub/emdb/structures/EMD-14232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14232 | HTTPS FTP |
-Validation report
| Summary document |  emd_14232_validation.pdf.gz emd_14232_validation.pdf.gz | 804.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14232_full_validation.pdf.gz emd_14232_full_validation.pdf.gz | 804.3 KB | Display | |
| Data in XML |  emd_14232_validation.xml.gz emd_14232_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_14232_validation.cif.gz emd_14232_validation.cif.gz | 28.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14232 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14232 | HTTPS FTP |
-Related structure data
| Related structure data |  7r16MC  7r0zC  7r10C  7r11C  7r12C  7r13C  7r14C  7r15C  7r17C  7r18C  7r19C  7r1aC  7r1bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14232.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14232.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
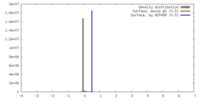
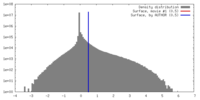
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_14232_msk_1.map emd_14232_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
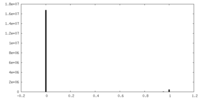
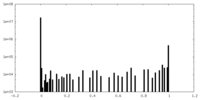
| Density Histograms |
-Half map: #2
| File | emd_14232_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14232_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Beta Variant SARS-CoV-2 Spike with 1 Erect RBD
| Entire | Name: Beta Variant SARS-CoV-2 Spike with 1 Erect RBD |
|---|---|
| Components |
|
-Supramolecule #1: Beta Variant SARS-CoV-2 Spike with 1 Erect RBD
| Supramolecule | Name: Beta Variant SARS-CoV-2 Spike with 1 Erect RBD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 420 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.864469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GMFVFLVLLP LVSSQCVNFT TRTQLPPAYT NSFTRGVYYP DKVFRSSVLH STQDLFLPF FSNVTWFHAI HVSGTNGTKR FANPVLPFND GVYFASTEKS NIIRGWIFGT TLDSKTQSLL IVNNATNVVI K VCEFQFCN ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GMFVFLVLLP LVSSQCVNFT TRTQLPPAYT NSFTRGVYYP DKVFRSSVLH STQDLFLPF FSNVTWFHAI HVSGTNGTKR FANPVLPFND GVYFASTEKS NIIRGWIFGT TLDSKTQSLL IVNNATNVVI K VCEFQFCN DPFLGVYYHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM DLEGKQGNFK NLREFVFKNI DGYFKIYSKH TP INLVRGL PQGFSALEPL VDLPIGINIT RFQTLHISYL TPGDSSSGWT AGAAAYYVGY LQPRTFLLKY NENGTITDAV DCA LDPLSE TKCTLKSFTV EKGIYQTSNF RVQPTESIVR FPNITNLCPF GEVFNATRFA SVYAWNRKRI SNCVADYSVL YNSA SFSTF KCYGVSPTKL NDLCFTNVYA DSFVIRGDEV RQIAPGQTGN IADYNYKLPD DFTGCVIAWN SNNLDSKVGG NYNYL YRLF RKSNLKPFER DISTEIYQAG STPCNGVKGF NCYFPLQSYG FQPTYGVGYQ PYRVVVLSFE LLHAPATVCG PKKSTN LVK NKCVNFNFNG LTGTGVLTES NKKFLPFQQF GRDIADTTDA VRDPQTLEIL DITPCSFGGV SVITPGTNTS NQVAVLY QG VNCTEVPVAI HADQLTPTWR VYSTGSNVFQ TRAGCLIGAE HVNNSYECDI PIGAGICASY QTQTNSPSRA SSVASQSI I AYTMSLGVEN SVAYSNNSIA IPTNFTISVT TEILPVSMTK TSVDCTMYIC GDSTECSNLL LQYGSFCTQL NRALTGIAV EQDKNTQEVF AQVKQIYKTP PIKDFGGFNF SQILPDPSKP SKRSFIEDLL FNKVTLADAG FIKQYGDCLG DIAARDLICA QKFNGLTVL PPLLTDEMIA QYTSALLAGT ITSGWTFGAG AALQIPFAMQ MAYRFNGIGV TQNVLYENQK LIANQFNSAI G KIQDSLSS TASALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QL IRAAEIR ASANLAATKM SECVLGQSKR VDFCGKGYHL MSFPQSAPHG VVFLHVTYVP AQEKNFTTAP AICHDGKAHF PRE GVFVSN GTHWFVTQRN FYEPQIITTD NTFVSGNCDV VIGIVNNTVY DPLQPELDSF KEELDKYFKN HTSPDVDLGD ISGI NASVV NIQKEIDRLN EVAKNLNESL IDLQELGKYE QSGRENLYFQ GGGGSGYIPE APRDGQAYVR KDGEWVLLST FLGHH HHHH UniProtKB: Spike glycoprotein |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 28 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)