[English] 日本語
 Yorodumi
Yorodumi- EMDB-1413: Structural aspects of RbfA action during small ribosomal subunit ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1413 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural aspects of RbfA action during small ribosomal subunit assembly. | |||||||||
 Map data Map data | Cryo-EM map of Thermus thermophilus 30S subunit and RbfA complex. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal small subunit binding / maturation of SSU-rRNA / small ribosomal subunit / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / translation / response to antibiotic / cytosol Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.5 Å | |||||||||
 Authors Authors | Datta PP / Wilson DN / Kawazoe M / Swami NK / Kaminishi T / Sharma MR / Booth TM / Takemoto C / Fucini P / Yokoyama S / Agrawal RK | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2007 Journal: Mol Cell / Year: 2007Title: Structural aspects of RbfA action during small ribosomal subunit assembly. Authors: Partha P Datta / Daniel N Wilson / Masahito Kawazoe / Neil K Swami / Tatsuya Kaminishi / Manjuli R Sharma / Timothy M Booth / Chie Takemoto / Paola Fucini / Shigeyuki Yokoyama / Rajendra K Agrawal /  Abstract: Ribosome binding factor A (RbfA) is a bacterial cold shock response protein, required for an efficient processing of the 5' end of the 16S ribosomal RNA (rRNA) during assembly of the small (30S) ...Ribosome binding factor A (RbfA) is a bacterial cold shock response protein, required for an efficient processing of the 5' end of the 16S ribosomal RNA (rRNA) during assembly of the small (30S) ribosomal subunit. Here we present a crystal structure of Thermus thermophilus (Tth) RbfA and a three-dimensional cryo-electron microscopic (EM) map of the Tth 30S*RbfA complex. RbfA binds to the 30S subunit in a position overlapping the binding sites of the A and P site tRNAs, and RbfA's functionally important C terminus extends toward the 5' end of the 16S rRNA. In the presence of RbfA, a portion of the 16S rRNA encompassing helix 44, which is known to be directly involved in mRNA decoding and tRNA binding, is displaced. These results shed light on the role played by RbfA during maturation of the 30S subunit, and also indicate how RbfA provides cells with a translational advantage under conditions of cold shock. | |||||||||
| History |
|
- Structure visualization
Structure visualization
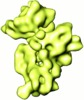
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1413.map.gz emd_1413.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1413-v30.xml emd-1413-v30.xml emd-1413.xml emd-1413.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1413.gif 1413.gif | 68.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1413 http://ftp.pdbj.org/pub/emdb/structures/EMD-1413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1413 | HTTPS FTP |
-Validation report
| Summary document |  emd_1413_validation.pdf.gz emd_1413_validation.pdf.gz | 294.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1413_full_validation.pdf.gz emd_1413_full_validation.pdf.gz | 293.8 KB | Display | |
| Data in XML |  emd_1413_validation.xml.gz emd_1413_validation.xml.gz | 5.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1413 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1413 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1413 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1413 | HTTPS FTP |
-Related structure data
| Related structure data |  2r1cMC  2r1gMC  2dyjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1413.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1413.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Thermus thermophilus 30S subunit and RbfA complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
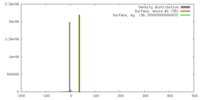
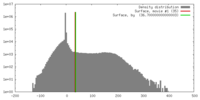
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of ribosomal subunit 30S and RbfA
| Entire | Name: Complex of ribosomal subunit 30S and RbfA |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of ribosomal subunit 30S and RbfA
| Supramolecule | Name: Complex of ribosomal subunit 30S and RbfA / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 910.9 KDa |
-Supramolecule #1: 30S small ribosomal subunit
| Supramolecule | Name: 30S small ribosomal subunit / type: complex / ID: 1 / Name.synonym: 30S / Details: Thermus thermophilus / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: SSU 30S |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
| Molecular weight | Experimental: 900 KDa |
-Macromolecule #1: Ribosome binding factor A
| Macromolecule | Name: Ribosome binding factor A / type: protein_or_peptide / ID: 1 / Name.synonym: RbfA / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Experimental: 10.9 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Details: 20mM Hepes-KOH (pH7.8), 10mM Mg(OAc)2, 200 mM NH4Cl, 65 mM KCL. |
|---|---|
| Grid | Details: Quantifoil 300 mesh Copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Wadworth Center own cryo-plunger fabricated on-site Method: 5ul of specimen was applied to the grid, then blotted using Whatman number 1 filter paper for 2 to 4 seconds, then plunged. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: Objective lends astigmatism corrected at 210x magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Number real images: 131 / Average electron dose: 20 e/Å2 / Bits/pixel: 12 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50760 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Oxford cryo-transfer holder / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 12.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 61207 |
|---|
-Atomic model buiding 1
| Software | Name: O |
|---|---|
| Details | Protocol: Rigid Body. The domains were fitted independently using the program O |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-2r1c:  PDB-2r1g: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)