+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inward-facing NPA bound form of auxin transporter PIN8 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Auxin transport / AEC family / BART superfamily / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationauxin export across the plasma membrane / auxin efflux transmembrane transporter activity / pollen development / auxin-activated signaling pathway / cell periphery / endoplasmic reticulum membrane / endoplasmic reticulum / protein homodimerization activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
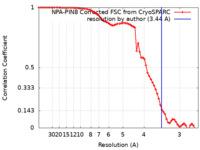
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Ung KL / Winkler MBL | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structures and mechanism of the plant PIN-FORMED auxin transporter. Authors: Kien Lam Ung / Mikael Winkler / Lukas Schulz / Martina Kolb / Dorina P Janacek / Emil Dedic / David L Stokes / Ulrich Z Hammes / Bjørn Panyella Pedersen /    Abstract: Auxins are hormones that have central roles and control nearly all aspects of growth and development in plants. The proteins in the PIN-FORMED (PIN) family (also known as the auxin efflux carrier ...Auxins are hormones that have central roles and control nearly all aspects of growth and development in plants. The proteins in the PIN-FORMED (PIN) family (also known as the auxin efflux carrier family) are key participants in this process and control auxin export from the cytosol to the extracellular space. Owing to a lack of structural and biochemical data, the molecular mechanism of PIN-mediated auxin transport is not understood. Here we present biophysical analysis together with three structures of Arabidopsis thaliana PIN8: two outward-facing conformations with and without auxin, and one inward-facing conformation bound to the herbicide naphthylphthalamic acid. The structure forms a homodimer, with each monomer divided into a transport and scaffold domain with a clearly defined auxin binding site. Next to the binding site, a proline-proline crossover is a pivot point for structural changes associated with transport, which we show to be independent of proton and ion gradients and probably driven by the negative charge of the auxin. The structures and biochemical data reveal an elevator-type transport mechanism reminiscent of bile acid/sodium symporters, bicarbonate/sodium symporters and sodium/proton antiporters. Our results provide a comprehensive molecular model for auxin recognition and transport by PINs, link and expand on a well-known conceptual framework for transport, and explain a central mechanism of polar auxin transport, a core feature of plant physiology, growth and development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14117.map.gz emd_14117.map.gz | 32.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14117-v30.xml emd-14117-v30.xml emd-14117.xml emd-14117.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14117_fsc.xml emd_14117_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14117.png emd_14117.png | 137.6 KB | ||
| Masks |  emd_14117_msk_1.map emd_14117_msk_1.map | 34.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14117.cif.gz emd-14117.cif.gz | 6.7 KB | ||
| Others |  emd_14117_half_map_1.map.gz emd_14117_half_map_1.map.gz emd_14117_half_map_2.map.gz emd_14117_half_map_2.map.gz | 31.7 MB 31.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14117 http://ftp.pdbj.org/pub/emdb/structures/EMD-14117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14117 | HTTPS FTP |
-Validation report
| Summary document |  emd_14117_validation.pdf.gz emd_14117_validation.pdf.gz | 818.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14117_full_validation.pdf.gz emd_14117_full_validation.pdf.gz | 818.4 KB | Display | |
| Data in XML |  emd_14117_validation.xml.gz emd_14117_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_14117_validation.cif.gz emd_14117_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14117 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14117 | HTTPS FTP |
-Related structure data
| Related structure data |  7qpcMC  7qp9C  7qpaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14117.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14117.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.294 Å | ||||||||||||||||||||||||||||||||||||
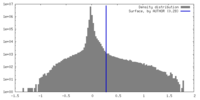
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14117_msk_1.map emd_14117_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
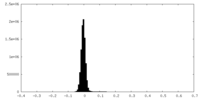
| Density Histograms |
-Half map: #2
| File | emd_14117_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
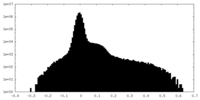
| Density Histograms |
-Half map: #1
| File | emd_14117_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
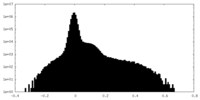
| Density Histograms |
- Sample components
Sample components
-Entire : NPA-form of PIN8
| Entire | Name: NPA-form of PIN8 |
|---|---|
| Components |
|
-Supramolecule #1: NPA-form of PIN8
| Supramolecule | Name: NPA-form of PIN8 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: dimer form of PIN8 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81 KDa |
-Macromolecule #1: Auxin efflux carrier component 8
| Macromolecule | Name: Auxin efflux carrier component 8 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.541039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGISWLDIYH VVSATVPLYV SMTLGFLSAR HLKLFSPEQC AGINKFVAKF SIPLLSFQII SENNPFKMSP KLILSDILQK FLVVVVLAM VLRFWHPTGG RGGKLGWVIT GLSISVLPNT LILGMPILSA IYGDEAASIL EQIVVLQSLI WYTILLFLFE L NAARALPS ...String: MGISWLDIYH VVSATVPLYV SMTLGFLSAR HLKLFSPEQC AGINKFVAKF SIPLLSFQII SENNPFKMSP KLILSDILQK FLVVVVLAM VLRFWHPTGG RGGKLGWVIT GLSISVLPNT LILGMPILSA IYGDEAASIL EQIVVLQSLI WYTILLFLFE L NAARALPS SGASLEHTGN DQEEANIEDE PKEEEDEEEV AIVRTRSVGT MKILLKAWRK LIINPNTYAT LIGIIWATLH FR LGWNLPE MIDKSIHLLS DGGLGMAMFS LGLFMASQSS IIACGTKMAI ITMLLKFVLG PALMIASAYC IRLKSTLFKV AIL QAALPQ GVVPFVFAKE YNLHPEIIST GVIFGMLIAL PTTLAYYFLL DLPGENLYFQ UniProtKB: Auxin efflux carrier component 8 |
-Macromolecule #2: 2-(naphthalen-1-ylcarbamoyl)benzoic acid
| Macromolecule | Name: 2-(naphthalen-1-ylcarbamoyl)benzoic acid / type: ligand / ID: 2 / Number of copies: 2 / Formula: E7O |
|---|---|
| Molecular weight | Theoretical: 291.301 Da |
| Chemical component information |  ChemComp-E7O: |
-Macromolecule #3: 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: DLP |
|---|---|
| Molecular weight | Theoretical: 782.082 Da |
| Chemical component information |  ChemComp-DLP: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 62 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Details: The grid was glow-discharge at 15 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Wait 4 seconds after sample loading, Blotting time 4 seconds with blotting force of -1 before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 14500 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)