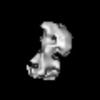[English] 日本語
 Yorodumi
Yorodumi- EMDB-1410: The EM structure of human DNA polymerase gamma reveals a localize... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1410 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The EM structure of human DNA polymerase gamma reveals a localized contact between the catalytic and accessory subunits. | |||||||||
 Map data Map data | This is a map of human mitochondrial DNA polymerase gamma | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 17.0 Å | |||||||||
 Authors Authors | Yakubovskaya E / Lukin M / Chen Z / Berriman J / Wall JS / Kobayashi R / Kisker C / Bogenhagen DF | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2007 Journal: EMBO J / Year: 2007Title: The EM structure of human DNA polymerase gamma reveals a localized contact between the catalytic and accessory subunits. Authors: Elena Yakubovskaya / Mark Lukin / Zhixin Chen / John Berriman / Joseph S Wall / Ryuji Kobayashi / Caroline Kisker / Daniel F Bogenhagen /  Abstract: We used electron microscopy to examine the structure of human DNA pol gamma, the heterotrimeric mtDNA replicase implicated in certain mitochondrial diseases and aging models. Separate analysis of ...We used electron microscopy to examine the structure of human DNA pol gamma, the heterotrimeric mtDNA replicase implicated in certain mitochondrial diseases and aging models. Separate analysis of negatively stained preparations of the catalytic subunit, pol gammaA, and of the holoenzyme including a dimeric accessory factor, pol gammaB(2), permitted unambiguous identification of the position of the accessory factor within the holoenzyme. The model explains protection of a partial chymotryptic cleavage site after residue L(549) of pol gammaA upon binding of the accessory subunit. This interaction region is near residue 467 of pol gammaA, where a disease-related mutation has been reported to impair binding of the B subunit. One pol gammaB subunit dominates contacts with the catalytic subunit, while the second B subunit is largely exposed to solvent. A model for pol gamma is discussed that considers the effects of known mutations in the accessory subunit and the interaction of the enzyme with DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1410.map.gz emd_1410.map.gz | 788.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1410-v30.xml emd-1410-v30.xml emd-1410.xml emd-1410.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1410.gif 1410.gif | 46.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1410 http://ftp.pdbj.org/pub/emdb/structures/EMD-1410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1410 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1410.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1410.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of human mitochondrial DNA polymerase gamma | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.488 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
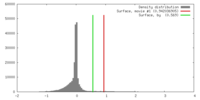
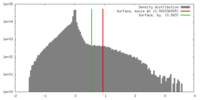
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human mitochondrial DNA polymerase gamma polymerase gamma
| Entire | Name: Human mitochondrial DNA polymerase gamma polymerase gamma |
|---|---|
| Components |
|
-Supramolecule #1000: Human mitochondrial DNA polymerase gamma polymerase gamma
| Supramolecule | Name: Human mitochondrial DNA polymerase gamma polymerase gamma type: sample / ID: 1000 / Oligomeric state: heterotrimer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 250 KDa / Theoretical: 250 KDa |
-Macromolecule #1: human mitochondrial DNA polymerase gamma, catalytic subunit
| Macromolecule | Name: human mitochondrial DNA polymerase gamma, catalytic subunit type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: mitochondria Homo sapiens (human) / synonym: Human / Location in cell: mitochondria |
| Molecular weight | Experimental: 145 KDa / Theoretical: 145 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: human mitochondrial DNA polymerase gamma, dimeric accessory subunit
| Macromolecule | Name: human mitochondrial DNA polymerase gamma, dimeric accessory subunit type: protein_or_peptide / ID: 2 / Details: dimer / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: mitochondria Homo sapiens (human) / synonym: Human / Location in cell: mitochondria |
| Molecular weight | Experimental: 110 KDa / Theoretical: 110 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Hepes, 150 mM KCl, 1 mM EDTA, 5 mM DTT buffer. |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein were stained with 2% uranyl acetate for 60 seconds. |
| Grid | Details: 300-mesh copper grids (Ted Pella) coated with carbon film that were glow-discharged for 10 s. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200T |
|---|---|
| Specialist optics | Energy filter - Name: FEI |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number real images: 48 / Average electron dose: 10 e/Å2 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 76000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.6 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.85 µm |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN HELIUM / Tilt angle min: 60 |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 8652 |
| Final two d classification | Number classes: 48 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)