[English] 日本語
 Yorodumi
Yorodumi- EMDB-14099: STRUCTURE OF THE HUMAN INNER KINETOCHORE CCAN COMPLEX AT 10A RESO... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF THE HUMAN INNER KINETOCHORE CCAN COMPLEX AT 10A RESOLUTION | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | INNER KINETOCHORE / CCAN / COMPLEX / DNA BINDING PROTEIN / CELL CYCLE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
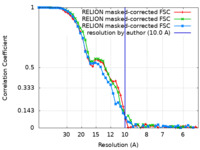
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Vetter IR / Pesenti M / Raisch T | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization. Authors: Marion E Pesenti / Tobias Raisch / Duccio Conti / Kai Walstein / Ingrid Hoffmann / Dorothee Vogt / Daniel Prumbaum / Ingrid R Vetter / Stefan Raunser / Andrea Musacchio /  Abstract: Centromeres are specialized chromosome loci that seed the kinetochore, a large protein complex that effects chromosome segregation. A 16-subunit complex, the constitutive centromere associated ...Centromeres are specialized chromosome loci that seed the kinetochore, a large protein complex that effects chromosome segregation. A 16-subunit complex, the constitutive centromere associated network (CCAN), connects between the specialized centromeric chromatin, marked by the histone H3 variant CENP-A, and the spindle-binding moiety of the kinetochore. Here, we report a cryo-electron microscopy structure of human CCAN. We highlight unique features such as the pseudo GTPase CENP-M and report how a crucial CENP-C motif binds the CENP-LN complex. The CCAN structure has implications for the mechanism of specific recognition of the CENP-A nucleosome. A model consistent with our structure depicts the CENP-C-bound nucleosome as connected to the CCAN through extended, flexible regions of CENP-C. An alternative model identifies both CENP-C and CENP-N as specificity determinants but requires CENP-N to bind CENP-A in a mode distinct from the classical nucleosome octamer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14099.map.gz emd_14099.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14099-v30.xml emd-14099-v30.xml emd-14099.xml emd-14099.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14099_fsc.xml emd_14099_fsc.xml emd_14099_fsc_2.xml emd_14099_fsc_2.xml emd_14099_fsc_3.xml emd_14099_fsc_3.xml | 4.7 KB 4.7 KB 4.7 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_14099.png emd_14099.png | 159.9 KB | ||
| Masks |  emd_14099_msk_1.map emd_14099_msk_1.map emd_14099_msk_2.map emd_14099_msk_2.map emd_14099_msk_3.map emd_14099_msk_3.map | 8 MB 8 MB 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14099.cif.gz emd-14099.cif.gz | 4.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14099 http://ftp.pdbj.org/pub/emdb/structures/EMD-14099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14099 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14099.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14099.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.72 Å | ||||||||||||||||||||||||||||||||||||
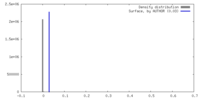
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14099_msk_1.map emd_14099_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
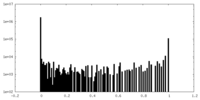
| Density Histograms |
-Mask #2
| File |  emd_14099_msk_2.map emd_14099_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
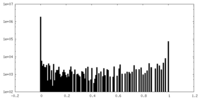
| Density Histograms |
-Mask #3
| File |  emd_14099_msk_3.map emd_14099_msk_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
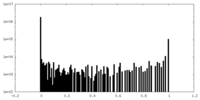
| Density Histograms |
- Sample components
Sample components
-Entire : human inner kinetochore CCAN complex
| Entire | Name: human inner kinetochore CCAN complex |
|---|---|
| Components |
|
-Supramolecule #1: human inner kinetochore CCAN complex
| Supramolecule | Name: human inner kinetochore CCAN complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#15 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 451 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Details: 0.0025% Triton |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Spherical aberration corrector: Krios with CS corrector / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 14 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.8 e/Å2 Details: 2678 images were collected in movie-mode with 60 frames per image on Gatan K3 in superresolution mode with 0.34 A pixel size / native pixel size 0.68A |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)