[English] 日本語
 Yorodumi
Yorodumi- EMDB-14083: 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationproteasome regulatory particle assembly / proteasome-activating activity / proteasome regulatory particle, base subcomplex / Cross-presentation of soluble exogenous antigens (endosomes) / TNFR2 non-canonical NF-kB pathway / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway ...proteasome regulatory particle assembly / proteasome-activating activity / proteasome regulatory particle, base subcomplex / Cross-presentation of soluble exogenous antigens (endosomes) / TNFR2 non-canonical NF-kB pathway / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / Neddylation / Orc1 removal from chromatin / MAPK6/MAPK4 signaling / regulation of protein catabolic process / proteasome storage granule / Antigen processing: Ubiquitination & Proteasome degradation / protein deubiquitination / Ub-specific processing proteases / enzyme regulator activity / Neutrophil degranulation / proteasome complex / protein-macromolecule adaptor activity / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / mitochondrial outer membrane / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / endoplasmic reticulum / ATP hydrolysis activity / ATP binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |    Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | ||||||||||||||||||
 Authors Authors | Hung KYS / Klumpe S / Eisele MR / Elsasser S / Geng TT / Cheng C / Joshi T / Rudack T / Sakata E / Finley D | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Allosteric control of Ubp6 and the proteasome via a bidirectional switch. Authors: Hung KYS / Klumpe S / Eisele MR / Elsasser S / Tian G / Sun S / Moroco JA / Cheng TC / Joshi T / Seibel T / Van Dalen D / Feng XH / Lu Y / Ovaa H / Engen JR / Lee BH / Rudack T / Sakata E / Finley D | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14083.map.gz emd_14083.map.gz | 55.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14083-v30.xml emd-14083-v30.xml emd-14083.xml emd-14083.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14083.png emd_14083.png | 116.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14083 http://ftp.pdbj.org/pub/emdb/structures/EMD-14083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14083 | HTTPS FTP |
-Related structure data
| Related structure data |  7qo4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14083.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14083.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.1 Å | ||||||||||||||||||||||||||||||||||||
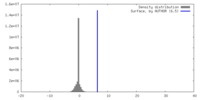
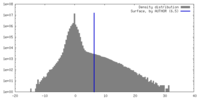
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : 26S proteasome WT-Ubp6-UbVS complex in the si state
+Supramolecule #1: 26S proteasome WT-Ubp6-UbVS complex in the si state
+Supramolecule #2: yeast WT 26S proteasome body2 (ATPase, Rpn1)
+Supramolecule #3: Ubp6-UbVS
+Macromolecule #1: 26S proteasome regulatory subunit RPN1
+Macromolecule #2: RPT1 isoform 1
+Macromolecule #3: RPT2 isoform 1
+Macromolecule #4: RPT3 isoform 1
+Macromolecule #5: RPT4 isoform 1
+Macromolecule #6: RPT5 isoform 1
+Macromolecule #7: RPT6 isoform 1
+Macromolecule #8: Ubiquitin carboxyl-terminal hydrolase
+Macromolecule #9: Polyubiquitin-B
+Macromolecule #10: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #11: MAGNESIUM ION
+Macromolecule #12: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 88243 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)