[English] 日本語
 Yorodumi
Yorodumi- EMDB-13967: Cryo-EM structure of the human mtLSU assembly intermediate upon M... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondria / Ribosome / Assembly / Methyltransferase / MRM2 / RNA modification | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of mitochondrial translation / mitochondrial large ribosomal subunit assembly / protein lipoylation / Complex I biogenesis / negative regulation of ribosome biogenesis / Mitochondrial Fatty Acid Beta-Oxidation / Protein lipoylation / positive regulation of mitochondrial translation / Respiratory electron transport / rRNA import into mitochondrion ...negative regulation of mitochondrial translation / mitochondrial large ribosomal subunit assembly / protein lipoylation / Complex I biogenesis / negative regulation of ribosome biogenesis / Mitochondrial Fatty Acid Beta-Oxidation / Protein lipoylation / positive regulation of mitochondrial translation / Respiratory electron transport / rRNA import into mitochondrion / mitochondrial translational elongation / Mitochondrial translation elongation / Mitochondrial translation termination / Mitochondrial translation initiation / mitochondrial [2Fe-2S] assembly complex / iron-sulfur cluster assembly complex / mitochondrial large ribosomal subunit / mitochondrial fission / mitochondrial large ribosomal subunit binding / mitochondrial ribosome / Hydrolases; Acting on ester bonds; Endoribonucleases producing 5'-phosphomonoesters / mitochondrial small ribosomal subunit / [2Fe-2S] cluster assembly / mitochondrial translation / iron-sulfur cluster assembly / acyl binding / acyl carrier activity / ribosomal large subunit binding / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / proton motive force-driven mitochondrial ATP synthesis / respiratory chain complex I / RNA processing / Mitochondrial protein degradation / cellular response to leukemia inhibitory factor / aerobic respiration / ribosomal large subunit biogenesis / fatty acid binding / mitochondrial membrane / fatty acid biosynthetic process / double-stranded RNA binding / 5S rRNA binding / small ribosomal subunit rRNA binding / endonuclease activity / mitochondrial inner membrane / negative regulation of translation / rRNA binding / nuclear body / ribosome / structural constituent of ribosome / mitochondrial matrix / protein domain specific binding / translation / ribonucleoprotein complex / nucleotide binding / mRNA binding / apoptotic process / calcium ion binding / mitochondrion / extracellular space / RNA binding / nucleoplasm / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
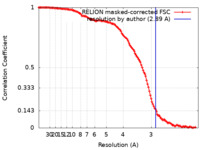
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Rebelo-Guiomar P / Pellegrino S / Dent KC / Warren AJ / Minczuk M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit. Authors: Pedro Rebelo-Guiomar / Simone Pellegrino / Kyle C Dent / Aldema Sas-Chen / Leonor Miller-Fleming / Caterina Garone / Lindsey Van Haute / Jack F Rogan / Adam Dinan / Andrew E Firth / Byron ...Authors: Pedro Rebelo-Guiomar / Simone Pellegrino / Kyle C Dent / Aldema Sas-Chen / Leonor Miller-Fleming / Caterina Garone / Lindsey Van Haute / Jack F Rogan / Adam Dinan / Andrew E Firth / Byron Andrews / Alexander J Whitworth / Schraga Schwartz / Alan J Warren / Michal Minczuk /    Abstract: Many cellular processes, including ribosome biogenesis, are regulated through post-transcriptional RNA modifications. Here, a genome-wide analysis of the human mitochondrial transcriptome shows that ...Many cellular processes, including ribosome biogenesis, are regulated through post-transcriptional RNA modifications. Here, a genome-wide analysis of the human mitochondrial transcriptome shows that 2'-O-methylation is limited to residues of the mitoribosomal large subunit (mtLSU) 16S mt-rRNA, introduced by MRM1, MRM2 and MRM3, with the modifications installed by the latter two proteins being interdependent. MRM2 controls mitochondrial respiration by regulating mitoribosome biogenesis. In its absence, mtLSU particles (visualized by cryo-EM at the resolution of 2.6 Å) present disordered RNA domains, partial occupancy of bL36m and bound MALSU1:L0R8F8:mtACP anti-association module, allowing five mtLSU biogenesis intermediates with different intersubunit interface configurations to be placed along the assembly pathway. However, mitoribosome biogenesis does not depend on the methyltransferase activity of MRM2. Disruption of the MRM2 Drosophila melanogaster orthologue leads to mitochondria-related developmental arrest. This work identifies a key checkpoint during mtLSU assembly, essential to maintain mitochondrial homeostasis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13967.map.gz emd_13967.map.gz | 139.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13967-v30.xml emd-13967-v30.xml emd-13967.xml emd-13967.xml | 62.3 KB 62.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13967_fsc.xml emd_13967_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13967.png emd_13967.png | 77.6 KB | ||
| Filedesc metadata |  emd-13967.cif.gz emd-13967.cif.gz | 13.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13967 http://ftp.pdbj.org/pub/emdb/structures/EMD-13967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13967 | HTTPS FTP |
-Related structure data
| Related structure data |  7qh7MC  7qh6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13967.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13967.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : Human mitochondrial ribosome large subunit
+Supramolecule #1: Human mitochondrial ribosome large subunit
+Macromolecule #1: 39S ribosomal protein L2, mitochondrial
+Macromolecule #2: 39S ribosomal protein L3, mitochondrial
+Macromolecule #3: 39S ribosomal protein L4, mitochondrial
+Macromolecule #4: 39S ribosomal protein L9, mitochondrial
+Macromolecule #5: 39S ribosomal protein L10, mitochondrial
+Macromolecule #6: 39S ribosomal protein L13, mitochondrial
+Macromolecule #7: 39S ribosomal protein L14, mitochondrial
+Macromolecule #8: 39S ribosomal protein L15, mitochondrial
+Macromolecule #9: 39S ribosomal protein L16, mitochondrial
+Macromolecule #10: 39S ribosomal protein L17, mitochondrial
+Macromolecule #11: 39S ribosomal protein L18, mitochondrial
+Macromolecule #12: 39S ribosomal protein L19, mitochondrial
+Macromolecule #13: 39S ribosomal protein L20, mitochondrial
+Macromolecule #14: 39S ribosomal protein L21, mitochondrial
+Macromolecule #15: 39S ribosomal protein L22, mitochondrial
+Macromolecule #16: 39S ribosomal protein L23, mitochondrial
+Macromolecule #17: 39S ribosomal protein L24, mitochondrial
+Macromolecule #18: 39S ribosomal protein L27, mitochondrial
+Macromolecule #19: 39S ribosomal protein L28, mitochondrial
+Macromolecule #20: 39S ribosomal protein L47, mitochondrial
+Macromolecule #21: 39S ribosomal protein L30, mitochondrial
+Macromolecule #22: 39S ribosomal protein L32, mitochondrial
+Macromolecule #23: 39S ribosomal protein L33, mitochondrial
+Macromolecule #24: 39S ribosomal protein L34, mitochondrial
+Macromolecule #25: 39S ribosomal protein L35, mitochondrial
+Macromolecule #26: 39S ribosomal protein L37, mitochondrial
+Macromolecule #27: 39S ribosomal protein L38, mitochondrial
+Macromolecule #28: 39S ribosomal protein L39, mitochondrial
+Macromolecule #29: 39S ribosomal protein L41, mitochondrial
+Macromolecule #30: 39S ribosomal protein L42, mitochondrial
+Macromolecule #31: 39S ribosomal protein L43, mitochondrial
+Macromolecule #32: 39S ribosomal protein L44, mitochondrial
+Macromolecule #33: 39S ribosomal protein L45, mitochondrial
+Macromolecule #34: 39S ribosomal protein L48, mitochondrial
+Macromolecule #35: 39S ribosomal protein L49, mitochondrial
+Macromolecule #36: 39S ribosomal protein L50, mitochondrial
+Macromolecule #37: 39S ribosomal protein L51, mitochondrial
+Macromolecule #38: 39S ribosomal protein L52, mitochondrial
+Macromolecule #39: Ribosomal protein 63, mitochondrial
+Macromolecule #40: Peptidyl-tRNA hydrolase ICT1, mitochondrial
+Macromolecule #41: Growth arrest and DNA damage-inducible proteins-interacting protein 1
+Macromolecule #42: 39S ribosomal protein S18a, mitochondrial
+Macromolecule #43: 39S ribosomal protein S30, mitochondrial
+Macromolecule #44: Mitochondrial assembly of ribosomal large subunit protein 1
+Macromolecule #45: MIEF1 upstream open reading frame protein
+Macromolecule #46: Acyl carrier protein, mitochondrial
+Macromolecule #49: 39S ribosomal protein L36, mitochondrial
+Macromolecule #47: 16S ribosomal RNA
+Macromolecule #48: mitochondrial tRNAVal
+Macromolecule #50: MAGNESIUM ION
+Macromolecule #51: ZINC ION
+Macromolecule #52: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 52.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)