+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S.c. Condensin peripheral Ycg1 subcomplex bound to DNA | |||||||||
 Map data Map data | cryoEM half map 1 of S.c. condensin peripheral Ycg1 complex bound to DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SMC-motor protein / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of meiotic DNA double-strand break formation / Condensation of Prometaphase Chromosomes / tRNA gene clustering / meiotic chromosome condensation / meiotic chromosome separation / condensin complex / rDNA chromatin condensation / synaptonemal complex assembly / mitotic chromosome condensation / chromosome condensation ...negative regulation of meiotic DNA double-strand break formation / Condensation of Prometaphase Chromosomes / tRNA gene clustering / meiotic chromosome condensation / meiotic chromosome separation / condensin complex / rDNA chromatin condensation / synaptonemal complex assembly / mitotic chromosome condensation / chromosome condensation / mitotic sister chromatid segregation / condensed chromosome / cell division / chromatin binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
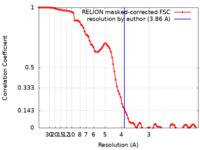
| Method | single particle reconstruction / cryo EM / Resolution: 3.86 Å | |||||||||
 Authors Authors | Lecomte L / Hassler M / Haering C / Eustermann S | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: A hold-and-feed mechanism drives directional DNA loop extrusion by condensin. Authors: Indra A Shaltiel / Sumanjit Datta / Léa Lecomte / Markus Hassler / Marc Kschonsak / Sol Bravo / Catherine Stober / Jenny Ormanns / Sebastian Eustermann / Christian H Haering /  Abstract: Structural maintenance of chromosomes (SMC) protein complexes structure genomes by extruding DNA loops, but the molecular mechanism that underlies their activity has remained unknown. We show that ...Structural maintenance of chromosomes (SMC) protein complexes structure genomes by extruding DNA loops, but the molecular mechanism that underlies their activity has remained unknown. We show that the active condensin complex entraps the bases of a DNA loop transiently in two separate chambers. Single-molecule imaging and cryo-electron microscopy suggest a putative power-stroke movement at the first chamber that feeds DNA into the SMC-kleisin ring upon adenosine triphosphate binding, whereas the second chamber holds on upstream of the same DNA double helix. Unlocking the strict separation of "motor" and "anchor" chambers turns condensin from a one-sided into a bidirectional DNA loop extruder. We conclude that the orientation of two topologically bound DNA segments during the SMC reaction cycle determines the directionality of DNA loop extrusion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13950.map.gz emd_13950.map.gz | 57 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13950-v30.xml emd-13950-v30.xml emd-13950.xml emd-13950.xml | 29.1 KB 29.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13950_fsc.xml emd_13950_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13950.png emd_13950.png | 63.7 KB | ||
| Masks |  emd_13950_msk_1.map emd_13950_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13950.cif.gz emd-13950.cif.gz | 8 KB | ||
| Others |  emd_13950_additional_1.map.gz emd_13950_additional_1.map.gz emd_13950_additional_2.map.gz emd_13950_additional_2.map.gz emd_13950_half_map_1.map.gz emd_13950_half_map_1.map.gz emd_13950_half_map_2.map.gz emd_13950_half_map_2.map.gz | 40.4 MB 49.6 MB 49.6 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13950 http://ftp.pdbj.org/pub/emdb/structures/EMD-13950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13950 | HTTPS FTP |
-Validation report
| Summary document |  emd_13950_validation.pdf.gz emd_13950_validation.pdf.gz | 913.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13950_full_validation.pdf.gz emd_13950_full_validation.pdf.gz | 913.4 KB | Display | |
| Data in XML |  emd_13950_validation.xml.gz emd_13950_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_13950_validation.cif.gz emd_13950_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13950 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13950 | HTTPS FTP |
-Related structure data
| Related structure data |  7qfwMC  7qenC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13950.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13950.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM half map 1 of S.c. condensin peripheral Ycg1 complex bound to DNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13950_msk_1.map emd_13950_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: cryoEM map of S.c. condensin peripheral Ycg1 complex...
| File | emd_13950_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM map of S.c. condensin peripheral Ycg1 complex bound to DNA local-resolution filtered and postprocessed cryo map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened cryoEM map of S.c. condensin peripheral Ycg1...
| File | emd_13950_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened cryoEM map of S.c. condensin peripheral Ycg1 complex bound to DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: cryoEM half map 2 of S.c. condensin peripheral...
| File | emd_13950_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM half map 2 of S.c. condensin peripheral Ycg1 complex bound to DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: cryoEM half map 2 of S.c. condensin peripheral...
| File | emd_13950_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM half map 2 of S.c. condensin peripheral Ycg1 complex bound to DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : S.c. Condensin peripheral Ycg1 complex bound to DNA
| Entire | Name: S.c. Condensin peripheral Ycg1 complex bound to DNA |
|---|---|
| Components |
|
-Supramolecule #1: S.c. Condensin peripheral Ycg1 complex bound to DNA
| Supramolecule | Name: S.c. Condensin peripheral Ycg1 complex bound to DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 707 KDa |
-Macromolecule #1: Condensin complex subunit 3
| Macromolecule | Name: Condensin complex subunit 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 117.981 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQDPDGIDIN TKIFNSVAEV FQKAQGSYAG HRKHIAVLKK IQSKAVEQGY EDAFNFWFDK LVTKILPLKK NEIIGDRIVK LVAAFIASL ERELILAKKQ NYKLTNDEEG IFSRFVDQFI RHVLRGVESP DKNVRFRVLQ LLAVIMDNIG EIDESLFNLL I LSLNKRIY ...String: MQDPDGIDIN TKIFNSVAEV FQKAQGSYAG HRKHIAVLKK IQSKAVEQGY EDAFNFWFDK LVTKILPLKK NEIIGDRIVK LVAAFIASL ERELILAKKQ NYKLTNDEEG IFSRFVDQFI RHVLRGVESP DKNVRFRVLQ LLAVIMDNIG EIDESLFNLL I LSLNKRIY DREPTVRIQA VFCLTKFQDE EQTEHLTELS DNEENFEATR TLVASIQNDP SAEVRRAAML NLINDNNTRP YI LERARDV NIVNRRLVYS RILKSMGRKC FDDIEPHIFD QLIEWGLEDR ELSVRNACKR LIAHDWLNAL DGDLIELLEK LDV SRSSVC VKAIEALFQS RPDILSKIKF PESIWKDFTV EIAFLFRAIY LYCLDNNITE MLEENFPEAS KLSEHLNHYI LLRY HHNDI SNDSQSHFDY NTLEFIIEQL SIAAERYDYS DEVGRRSMLT VVRNMLALTT LSEPLIKIGI RVMKSLSINE KDFVT MAIE IINDIRDDDI EKQEQEEKIK SKKINRRNET SVDEEDENGT HNDEVNEDEE DDNISSFHSA VENLVQGNGN VSESDI INN LPPEKEASSA TIVLCLTRSS YMLELVNTPL TENILIASLM DTLITPAVRN TAPNIRELGV KNLGLCCLLD VKLAIDN MY ILGMCVSKGN ASLKYIALQV IVDIFSVHGN TVVDGEGKVD SISLHKIFYK VLKNNGLPEC QVIAAEGLCK LFLADVFT D DDLFETLVLS YFSPINSSNE ALVQAFAFCI PVYCFSHPAH QQRMSRTAAD ILLRLCVLWD DLQSSVIPEV DREAMLKPN IIFQQLLFWT DPRNLVNQTG STKKDTVQLT FLIDVLKIYA QIEKKEIKKM IITNINAIFL SSEQDYSTLK ELLEYSDDIA ENDNLDNVS KNALDKLRNN LNSLIEEINE RSETQTKDEN NTANDQYSSI LGNSFNKSSN DTIEHAADIT DGNNTELTKT T VNISAVDN TTEQSNSRKR TRSEAEQIDT SKNLENMSIQ DTSTVAKNVS FVLPDEKSDA MSIDEEDKDS ESFSEVC UniProtKB: Condensin complex subunit 3 |
-Macromolecule #2: Condensin complex subunit 2
| Macromolecule | Name: Condensin complex subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.721219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTQLRYENN DDDERVEYNL FTNRSTMMAN FEEWIKMATD NKINSRNSWN FALIDYFYDL DVLKDGENNI NFQKASATLD GCIKIYSSR VDSVTTETGK LLSGLAQRKT NGASNGDDSN GGNGEGLGGD SDEANIEIDP LTGMPISNDP DVNNTRRRVY N RVLETTLV ...String: MTTQLRYENN DDDERVEYNL FTNRSTMMAN FEEWIKMATD NKINSRNSWN FALIDYFYDL DVLKDGENNI NFQKASATLD GCIKIYSSR VDSVTTETGK LLSGLAQRKT NGASNGDDSN GGNGEGLGGD SDEANIEIDP LTGMPISNDP DVNNTRRRVY N RVLETTLV EFETIKMKEL DQELIIDPLF KKALVDFDEG GAKSLLLNTL NIDNTARVIF DASIKDTQNV GQGKLQRKEE EL IERDSLV DDENEPSQSL ISTRNDSTVN DSVISAPSME DEILSLGMDF IKFDQIAVCE ISGSIEQLRN VVEDINQAKD FIE NVNNRF DNFLTEEELQ AAVPDNAEDD SDGFDMGMQQ ELCYPDENHD NTSHDEQDDD NVNSTTGSIF EKDLMAYFDE NLNR NWRGR EHWKVRNFKK ANLVNKESDL LEETRTTIGD TTDKNTTDDK SMDTKKKHKQ KKVLEIDFFK TDDSFEDKVF ASKGR TKID MPIKNRKNDT HYLLPDDFHF STDRITRLFI KPAQKMSLFS HRKHTRGDVS SGLFEKSTVS ANHSNNDIPT IADEHF WAD NYERKEQEEK EKEQSKEVGD VVGGALDNPF EDDMDGVDFN QAFEGTDDNE EASVKLDLQD DEDHKFPIRE NKVTYSR VS KKVDVRRLKK NVWRSINNLI QEHDSRKNRE QSSNDSETHT EDESTKELKF SDIIQGISKM YSDDTLKDIS TSFCFICL L HLANEHGLQI THTENYNDLI VNYEDLATTQ AASLVGGGHH RPHHGGHHHH HHGGRIFYPY DVPDYAGYPY DVPDYAGSY PYDVPNYAAG H UniProtKB: Condensin complex subunit 2 |
-Macromolecule #3: Synthetic DNA ligand
| Macromolecule | Name: Synthetic DNA ligand / type: dna / ID: 3 Details: 50bp synthetic DNA ligand with the sequence: 5'-GTTGACAGTG TCGCAACCTG CACAGGCAAG CTGCTGAGTC TGGTGTAGAC-3' The DNA ligand was modelled as poly(dA) as the register could not be determined by cryoEM density. Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.615376 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA) |
-Macromolecule #4: synthetic DNA ligand
| Macromolecule | Name: synthetic DNA ligand / type: dna / ID: 4 Details: 50bp synthetic DNA ligand with the sequence: 5'-GTCTACACCAGACTCAGCAGCTTGCCTGTGCAGGTTGCGACACTGTCAAC-3' The DNA ligand was modelled as poly(dT) as the register could not be determined by cryoEM density. Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.164683 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.707 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 6544 / Average exposure time: 8.0 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)