+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lanreotide nanotube | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Lanreotide / nanotube / assembly / HORMONE | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
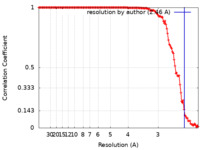
| Method | helical reconstruction / cryo EM / Resolution: 2.46 Å | ||||||||||||
 Authors Authors | Pieri L / Wang F | ||||||||||||
| Funding support |  France, France,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Atomic structure of Lanreotide nanotubes revealed by cryo-EM. Authors: Laura Pieri / Fengbin Wang / Ana-Andreea Arteni / Matthijn Vos / Jean-Marie Winter / Marie-Hélène Le Du / Franck Artzner / Frédéric Gobeaux / Pierre Legrand / Yves Boulard / Stéphane ...Authors: Laura Pieri / Fengbin Wang / Ana-Andreea Arteni / Matthijn Vos / Jean-Marie Winter / Marie-Hélène Le Du / Franck Artzner / Frédéric Gobeaux / Pierre Legrand / Yves Boulard / Stéphane Bressanelli / Edward H Egelman / Maité Paternostre /   Abstract: Functional and versatile nano- and microassemblies formed by biological molecules are found at all levels of life, from cell organelles to full organisms. Understanding the chemical and ...Functional and versatile nano- and microassemblies formed by biological molecules are found at all levels of life, from cell organelles to full organisms. Understanding the chemical and physicochemical determinants guiding the formation of these assemblies is crucial not only to understand the biological processes they carry out but also to mimic nature. Among the synthetic peptides forming well-defined nanostructures, the octapeptide Lanreotide has been considered one of the best characterized, in terms of both the atomic structure and its self-assembly process. In the present work, we determined the atomic structure of Lanreotide nanotubes at 2.5-Å resolution by cryoelectron microscopy (cryo-EM). Surprisingly, the asymmetric unit in the nanotube contains eight copies of the peptide, forming two tetramers. There are thus eight different environments for the peptide, and eight different conformations in the nanotube. The structure built from the cryo-EM map is strikingly different from the molecular model, largely based on X-ray fiber diffraction, proposed 20 y ago. Comparison of the nanotube with a crystal structure at 0.83-Å resolution of a Lanreotide derivative highlights the polymorphism for this peptide family. This work shows once again that higher-order assemblies formed by even well-characterized small peptides are very difficult to predict. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13830.map.gz emd_13830.map.gz | 69.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13830-v30.xml emd-13830-v30.xml emd-13830.xml emd-13830.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13830_fsc.xml emd_13830_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_13830.png emd_13830.png | 169.4 KB | ||
| Filedesc metadata |  emd-13830.cif.gz emd-13830.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13830 http://ftp.pdbj.org/pub/emdb/structures/EMD-13830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13830 | HTTPS FTP |
-Related structure data
| Related structure data |  7q5aMC  7q5gC M: atomic model generated by this map C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10873 (Title: Atomic structure of Lanreotide nanotubes revealed by cryo-EM EMPIAR-10873 (Title: Atomic structure of Lanreotide nanotubes revealed by cryo-EMData size: 362.5 Data #1: Lanreotide nanotube, subset of the final particles [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13830.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13830.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1102 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Lanreotide
| Entire | Name: Lanreotide |
|---|---|
| Components |
|
-Supramolecule #1: Lanreotide
| Supramolecule | Name: Lanreotide / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.096 kDa/nm |
-Macromolecule #1: Lanreotide
| Macromolecule | Name: Lanreotide / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.098339 KDa |
| Sequence | String: (4J2)CY(DTR)KVCT(NH2) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL |
|---|---|
| Buffer | pH: 6 / Component - Concentration: 40.0 mg/ml / Component - Name: Pure water |
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.039 kPa / Details: Current intensity 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 30 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: No corrector / Chromatic aberration corrector: No corrector / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV / Details: Bioquantum + K3 camera |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 19497 / Average exposure time: 1.69 sec. / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-7q5a: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)