+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
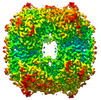
| Title | CryoEM structure of mammalian acylaminoacyl-peptidase | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | acylaminoacyl-peptidase / tetramer / aclypeptide-hydrolase / oxidized protein hydrolase / serine-protease / HYDROLASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationacylaminoacyl-peptidase / omega peptidase activity / serine-type endopeptidase activity / proteolysis / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | ||||||||||||||||||
 Authors Authors | Kiss-Szeman AJ / Harmat V | ||||||||||||||||||
| Funding support |  Hungary, 5 items Hungary, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Chem Sci / Year: 2022 Journal: Chem Sci / Year: 2022Title: Cryo-EM structure of acylpeptide hydrolase reveals substrate selection by multimerization and a multi-state serine-protease triad. Authors: Anna J Kiss-Szemán / Pál Stráner / Imre Jákli / Naoki Hosogi / Veronika Harmat / Dóra K Menyhárd / András Perczel /   Abstract: The first structure of tetrameric mammalian acylaminoacyl peptidase, an enzyme that functions as an upstream regulator of the proteasome through the removal of terminal -acetylated residues from its ...The first structure of tetrameric mammalian acylaminoacyl peptidase, an enzyme that functions as an upstream regulator of the proteasome through the removal of terminal -acetylated residues from its protein substrates, was determined by cryo-EM and further elucidated by MD simulations. Self-association results in a toroid-shaped quaternary structure, guided by an amyloidogenic β-edge and unique inserts. With a Pro introduced into its central β-sheet, sufficient conformational freedom is awarded to the segment containing the catalytic Ser587 that the serine protease catalytic triad alternates between active and latent states. Active site flexibility suggests that the dual function of catalysis and substrate selection are fulfilled by a novel mechanism: substrate entrance is regulated by flexible loops creating a double-gated channel system, while binding of the substrate to the active site is required for stabilization of the catalytic apparatus - as a second filter before hydrolysis. The structure not only underlines that within the family of S9 proteases homo-multimerization acts as a crucial tool for substrate selection, but it will also allow drug design targeting of the ubiquitin-proteasome system. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13691.map.gz emd_13691.map.gz | 11.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13691-v30.xml emd-13691-v30.xml emd-13691.xml emd-13691.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13691.png emd_13691.png | 224.1 KB | ||
| Filedesc metadata |  emd-13691.cif.gz emd-13691.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13691 http://ftp.pdbj.org/pub/emdb/structures/EMD-13691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13691 | HTTPS FTP |
-Validation report
| Summary document |  emd_13691_validation.pdf.gz emd_13691_validation.pdf.gz | 390.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13691_full_validation.pdf.gz emd_13691_full_validation.pdf.gz | 390.4 KB | Display | |
| Data in XML |  emd_13691_validation.xml.gz emd_13691_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_13691_validation.cif.gz emd_13691_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13691 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13691 | HTTPS FTP |
-Related structure data
| Related structure data |  7px8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13691.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13691.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
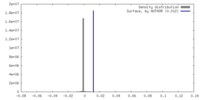
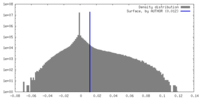
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Homotetramer of acylaminoacyl-peptidase
| Entire | Name: Homotetramer of acylaminoacyl-peptidase |
|---|---|
| Components |
|
-Supramolecule #1: Homotetramer of acylaminoacyl-peptidase
| Supramolecule | Name: Homotetramer of acylaminoacyl-peptidase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Acylamino-acid-releasing enzyme
| Macromolecule | Name: Acylamino-acid-releasing enzyme / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.324391 KDa |
| Sequence | String: MERQVLLSEP EEAAALYRGL SRQPALSAAC LGPEVTTQYG GRYRTVHTEW TQRDLERMEN IRFCRQYLVF HDGDSVVFAG PAGNSVETR GELLSRESPS GTMKAVLRKA GGTGTAEEKQ FLEVWEKNRK LKSFNLSALE KHGPVYEDDC FGCLSWSHSE T HLLYVADK ...String: MERQVLLSEP EEAAALYRGL SRQPALSAAC LGPEVTTQYG GRYRTVHTEW TQRDLERMEN IRFCRQYLVF HDGDSVVFAG PAGNSVETR GELLSRESPS GTMKAVLRKA GGTGTAEEKQ FLEVWEKNRK LKSFNLSALE KHGPVYEDDC FGCLSWSHSE T HLLYVADK KRPKAESFFQ TKALDVTGSD DEMARTKKPD QAIKGDQFLF YEDWGENMVS KSTPVLCVLD IESGNISVLE GV PESVSPG QAFWAPGDTG VVFVGWWHEP FRLGIRFCTN RRSALYYVDL TGGKCELLSD ESVAVTSPRL SPDQCRIVYL RFP SLVPHQ QCGQLCLYDW YTRVTSVVVD IVPRQLGEDF SGIYCSLLPL GCWSADSQRV VFDSPQRSRQ DLFAVDTQMG SVTS LTAGG SGGSWKLLTI DRDLMVVQFS TPSVPPSLKV GFLPPAGKEQ AVSWVSLEEA EPFPDISWSI RVLQPPPQQE HVQYA GLDF EAILLQPSNS PEKTQVPMVV MPHGGPHSSF VTAWMLFPAM LCKMGFAVLL VNYRGSTGFG QDSILSLPGN VGHQDV KDV QFAVEQVLQE EHFDAGRVAL MGGSHGGFLS CHLIGQYPET YSACVVRNPV INIASMMGST DIPDWCMVEA GFSYSSD CL PDLSVWAAML DKSPIKYAPQ VKTPLLLMLG QEDRRVPFKQ GMEYYRVLKA RNVPVRLLLY PKSTHALSEV EVESDSFM N AVLWLCTHLG S UniProtKB: Acylamino-acid-releasing enzyme |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 10.0 mM / Component - Formula: C4H11NO3 / Component - Name: TRIS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.015 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 1157 / Average exposure time: 4.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)