[English] 日本語
 Yorodumi
Yorodumi- EMDB-13614: S. cerevisiae Atm1 in MSP1D1 nanodiscs with bound AMP-PNP and Mg2+ -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13614 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae Atm1 in MSP1D1 nanodiscs with bound AMP-PNP and Mg2+ | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationiron-sulfur cluster transmembrane transport / Mitochondrial ABC transporters / iron-sulfur cluster export from the mitochondrion / mitochondrial transmembrane transport / Translocases / iron-sulfur cluster assembly / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transmembrane transport / intracellular iron ion homeostasis ...iron-sulfur cluster transmembrane transport / Mitochondrial ABC transporters / iron-sulfur cluster export from the mitochondrion / mitochondrial transmembrane transport / Translocases / iron-sulfur cluster assembly / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transmembrane transport / intracellular iron ion homeostasis / mitochondrial inner membrane / ATP hydrolysis activity / mitochondrion / ATP binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
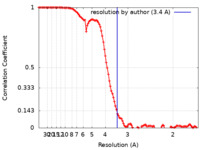
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Ellinghaus TL / Kuehlbrandt W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Conformational changes in the yeast mitochondrial ABC transporter Atm1 during the transport cycle. Authors: Thomas L Ellinghaus / Thomas Marcellino / Vasundara Srinivasan / Roland Lill / Werner Kühlbrandt /  Abstract: The mitochondrial inner membrane ABC transporter Atm1 exports an unknown substrate to the cytosol for iron-sulfur protein biogenesis, cellular iron regulation, and tRNA thio-modification. Mutations ...The mitochondrial inner membrane ABC transporter Atm1 exports an unknown substrate to the cytosol for iron-sulfur protein biogenesis, cellular iron regulation, and tRNA thio-modification. Mutations in the human relative ABCB7 cause the iron storage disease XLSA/A. We determined 3D structures of two complementary states of Atm1 in lipid nanodiscs by electron cryo-microscopy at 2.9- to 3.4-Å resolution. The inward-open structure resembled the known crystal structure of nucleotide-free apo-Atm1 closely. The occluded conformation with bound AMP-PNP-Mg showed a tight association of the two nucleotide-binding domains, a rearrangement of the C-terminal helices, and closure of the putative substrate-binding cavity in the homodimeric transporter. We identified a hydrophobic patch on the C-terminal helices of yeast Atm1, which is unique among type IV ABC transporters of known structure. Truncation mutants of yeast Atm1 suggest that the C-terminal helices stabilize the dimer, yet are not necessary for closure of the nucleotide-binding domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13614.map.gz emd_13614.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13614-v30.xml emd-13614-v30.xml emd-13614.xml emd-13614.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13614_fsc.xml emd_13614_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13614.png emd_13614.png | 202.8 KB | ||
| Filedesc metadata |  emd-13614.cif.gz emd-13614.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13614 http://ftp.pdbj.org/pub/emdb/structures/EMD-13614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13614 | HTTPS FTP |
-Validation report
| Summary document |  emd_13614_validation.pdf.gz emd_13614_validation.pdf.gz | 513.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13614_full_validation.pdf.gz emd_13614_full_validation.pdf.gz | 512.9 KB | Display | |
| Data in XML |  emd_13614_validation.xml.gz emd_13614_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_13614_validation.cif.gz emd_13614_validation.cif.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13614 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13614 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13614 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13614 | HTTPS FTP |
-Related structure data
| Related structure data |  7psmMC  7pslC  7psnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13614.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13614.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
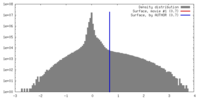
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Atm1 homodimer in MSP1D1 nanodiscs
| Entire | Name: Atm1 homodimer in MSP1D1 nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: Atm1 homodimer in MSP1D1 nanodiscs
| Supramolecule | Name: Atm1 homodimer in MSP1D1 nanodiscs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 137 KDa |
-Macromolecule #1: Iron-sulfur clusters transporter ATM1, mitochondrial
| Macromolecule | Name: Iron-sulfur clusters transporter ATM1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Translocases |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 68.392969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LKDLFRYIWP KGNNKVRIRV LIALGLLISA KILNVQVPFF FKQTIDSMNI AWDDPTVALP AAIGLTILCY GVARFGSVLF GELRNAVFA KVAQNAIRTV SLQTFQHLMK LDLGWHLSRQ TGGLTRAMDR GTKGISQVLT AMVFHIIPIS FEISVVCGIL T YQFGASFA ...String: LKDLFRYIWP KGNNKVRIRV LIALGLLISA KILNVQVPFF FKQTIDSMNI AWDDPTVALP AAIGLTILCY GVARFGSVLF GELRNAVFA KVAQNAIRTV SLQTFQHLMK LDLGWHLSRQ TGGLTRAMDR GTKGISQVLT AMVFHIIPIS FEISVVCGIL T YQFGASFA AITFSTMLLY SIFTIKTTAW RTHFRRDANK ADNKAASVAL DSLINFEAVK YFNNEKYLAD KYNGSLMNYR DS QIKVSQS LAFLNSGQNL IFTTALTAMM YMGCTGVIGG NLTVGDLVLI NQLVFQLSVP LNFLGSVYRD LKQSLIDMET LFK LRKNEV KIKNAERPLM LPENVPYDIT FENVTFGYHP DRKILKNASF TIPAGWKTAI VGSSGSGKST ILKLVFRFYD PESG RILIN GRDIKEYDID ALRKVIGVVP QDTPLFNDTI WENVKFGRID ATDEEVITVV EKAQLAPLIK KLPQGFDTIV GERGL MISG GEKQRLAIAR VLLKNARIMF FDEATSALDT HTEQALLRTI RDNFTSGSRT SVYIAHRLRT IADADKIIVL DNGRVR EEG KHLELLAMPG SLYRELWTIQ EDLDHLENEL KDQQELWSHP QFEK UniProtKB: Iron-sulfur clusters transporter ATM1, mitochondrial |
-Macromolecule #2: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANO...
| Macromolecule | Name: (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE type: ligand / ID: 4 / Number of copies: 2 / Formula: LOP |
|---|---|
| Molecular weight | Theoretical: 661.89 Da |
| Chemical component information |  ChemComp-LOP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)