[English] 日本語
 Yorodumi
Yorodumi- EMDB-13520: Cryo-EM structures of human fucosidase FucA1 reveal insight into ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Fucosidase / HYDROLASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglycolipid catabolic process / alpha-L-fucosidase / Reactions specific to the complex N-glycan synthesis pathway / alpha-L-fucosidase activity / fucose metabolic process / glycoside catabolic process / lysosomal lumen / azurophil granule lumen / lysosome / Neutrophil degranulation ...glycolipid catabolic process / alpha-L-fucosidase / Reactions specific to the complex N-glycan synthesis pathway / alpha-L-fucosidase activity / fucose metabolic process / glycoside catabolic process / lysosomal lumen / azurophil granule lumen / lysosome / Neutrophil degranulation / extracellular exosome / extracellular region / membrane / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.49 Å | |||||||||||||||
 Authors Authors | Armstrong Z / Meek RW | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis. Authors: Zachary Armstrong / Richard W Meek / Liang Wu / James N Blaza / Gideon J Davies /  Abstract: Enzymatic hydrolysis of α-L-fucose from fucosylated glycoconjugates is consequential in bacterial infections and the neurodegenerative lysosomal storage disorder fucosidosis. Understanding human α- ...Enzymatic hydrolysis of α-L-fucose from fucosylated glycoconjugates is consequential in bacterial infections and the neurodegenerative lysosomal storage disorder fucosidosis. Understanding human α-L-fucosidase catalysis, in an effort toward drug design, has been hindered by the absence of three-dimensional structural data for any animal fucosidase. Here, we have used cryoelectron microscopy (cryo-EM) to determine the structure of human lysosomal α-L-fucosidase (FucA1) in both an unliganded state and in complex with the inhibitor deoxyfuconojirimycin. These structures, determined at 2.49 Å resolution, reveal the homotetrameric structure of FucA1, the architecture of the catalytic center, and the location of both natural population variations and disease-causing mutations. Furthermore, this work has conclusively identified the hitherto contentious identity of the catalytic acid/base as aspartate-276, representing a shift from both the canonical glutamate acid/base residue and a previously proposed glutamate residue. These findings have furthered our understanding of how FucA1 functions in both health and disease. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13520.map.gz emd_13520.map.gz | 21.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13520-v30.xml emd-13520-v30.xml emd-13520.xml emd-13520.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13520.png emd_13520.png | 159.1 KB | ||
| Filedesc metadata |  emd-13520.cif.gz emd-13520.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13520 http://ftp.pdbj.org/pub/emdb/structures/EMD-13520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13520 | HTTPS FTP |
-Related structure data
| Related structure data |  7pm4MC  7plsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13520.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13520.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.656 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : FucA1 homotetramer in complex with deoxyfuconojirimycin
| Entire | Name: FucA1 homotetramer in complex with deoxyfuconojirimycin |
|---|---|
| Components |
|
-Supramolecule #1: FucA1 homotetramer in complex with deoxyfuconojirimycin
| Supramolecule | Name: FucA1 homotetramer in complex with deoxyfuconojirimycin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 225 kDa/nm |
-Macromolecule #1: Tissue alpha-L-fucosidase
| Macromolecule | Name: Tissue alpha-L-fucosidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: alpha-L-fucosidase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.525625 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GQPPRRYTPD WPSLDSRPLP AWFDEAKFGV FIHWGVFSVP AWGSEWFWWH WQGEGRPQYQ RFMRDNYPPG FSYADFGPQF TARFFHPEE WADLFQAAGA KYVVLTTKHH EGFTNWPSPV SWNWNSKDVG PHRDLVGELG TALRKRNIRY GLYHSLLEWF H PLYLLDKK ...String: GQPPRRYTPD WPSLDSRPLP AWFDEAKFGV FIHWGVFSVP AWGSEWFWWH WQGEGRPQYQ RFMRDNYPPG FSYADFGPQF TARFFHPEE WADLFQAAGA KYVVLTTKHH EGFTNWPSPV SWNWNSKDVG PHRDLVGELG TALRKRNIRY GLYHSLLEWF H PLYLLDKK NGFKTQHFVS AKTMPELYDL VNSYKPDLIW SDGEWECPDT YWNSTNFLSW LYNDSPVKDE VVVNDRWGQN CS CHHGGYY NCEDKFKPQS LPDHKWEMCT SIDKFSWGYR RDMALSDVTE ESEIISELVQ TVSLGGNYLL NIGPTKDGLI VPI FQERLL AVGKWLSING EAIYASKPWR VQWEKNTTSV WYTSKGSAVY AIFLHWPENG VLNLESPITT STTKITMLGI QGDL KWSTD PDKGLFISLP QLPPSAVPAE FAWTIKLTGV K UniProtKB: Tissue alpha-L-fucosidase |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL
| Macromolecule | Name: (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL / type: ligand / ID: 3 / Number of copies: 4 / Formula: DFU |
|---|---|
| Molecular weight | Theoretical: 147.172 Da |
| Chemical component information | 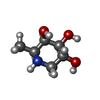 ChemComp-DFU: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 60 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 4.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 2 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D2 (2x2 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 2.49 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1.2) / Number images used: 98815 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)