[English] 日本語
 Yorodumi
Yorodumi- EMDB-13192: Structure of knob spiral in P. falciparum-infected erythrocytes -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13192 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of knob spiral in P. falciparum-infected erythrocytes | |||||||||
 Map data Map data | EM_map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
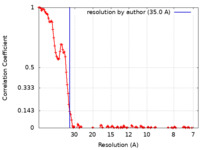
| Method | subtomogram averaging / cryo EM / Resolution: 35.0 Å | |||||||||
 Authors Authors | Chang S-YS / Kudryashev M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Microbiol / Year: 2022 Journal: Mol Microbiol / Year: 2022Title: KAHRP dynamically relocalizes to remodeled actin junctions and associates with knob spirals in Plasmodium falciparum-infected erythrocytes. Authors: Cecilia P Sanchez / Pintu Patra / Shih-Ying Scott Chang / Christos Karathanasis / Lukas Hanebutte / Nicole Kilian / Marek Cyrklaff / Mike Heilemann / Ulrich S Schwarz / Mikhail Kudryashev / Michael Lanzer /  Abstract: The knob-associated histidine-rich protein (KAHRP) plays a pivotal role in the pathophysiology of Plasmodium falciparum malaria by forming membrane protrusions in infected erythrocytes, which anchor ...The knob-associated histidine-rich protein (KAHRP) plays a pivotal role in the pathophysiology of Plasmodium falciparum malaria by forming membrane protrusions in infected erythrocytes, which anchor parasite-encoded adhesins to the membrane skeleton. The resulting sequestration of parasitized erythrocytes in the microvasculature leads to severe disease. Despite KAHRP being an important virulence factor, its physical location within the membrane skeleton is still debated, as is its function in knob formation. Here, we show by super-resolution microscopy that KAHRP initially associates with various skeletal components, including ankyrin bridges, but eventually colocalizes with remnant actin junctions. We further present a 35 Å map of the spiral scaffold underlying knobs and show that a KAHRP-targeting nanoprobe binds close to the spiral scaffold. Single-molecule localization microscopy detected ~60 KAHRP molecules/knob. We propose a dynamic model of KAHRP organization and a function of KAHRP in attaching other factors to the spiral scaffold. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13192.map.gz emd_13192.map.gz | 167.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13192-v30.xml emd-13192-v30.xml emd-13192.xml emd-13192.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13192_fsc.xml emd_13192_fsc.xml | 16.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13192.png emd_13192.png | 127.1 KB | ||
| Masks |  emd_13192_msk_1.map emd_13192_msk_1.map | 178 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13192 http://ftp.pdbj.org/pub/emdb/structures/EMD-13192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13192 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13192.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13192.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM_map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13192_msk_1.map emd_13192_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Knob spiral
| Entire | Name: Knob spiral |
|---|---|
| Components |
|
-Supramolecule #1: Knob spiral
| Supramolecule | Name: Knob spiral / type: complex / ID: 1 / Parent: 0 / Details: Knob spiral in P. falciparum-infected erythrocytes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS buffer |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK III |
| Details | P. falciparum-infected erythrocytes |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)