+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the hexameric 5S RNP from C. thermophilum | ||||||||||||
 Map data Map data | Hexameric 5S RNP from C. thermophilum | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | 5S RNP / Ribosome biogenesis / Symportin 1 / Rpf2-Rrs1 / RNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / protein import into nucleus / unfolded protein binding / ribosome biogenesis / ribosomal large subunit assembly / 5S rRNA binding / cytosolic large ribosomal subunit / rRNA binding / structural constituent of ribosome ...ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / protein import into nucleus / unfolded protein binding / ribosome biogenesis / ribosomal large subunit assembly / 5S rRNA binding / cytosolic large ribosomal subunit / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / nucleolus / nucleus Similarity search - Function | ||||||||||||
| Biological species |  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) / Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) /  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Castillo N / Thoms M | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways. Authors: Nestor Miguel Castillo Duque de Estrada / Matthias Thoms / Dirk Flemming / Henrik M Hammaren / Robert Buschauer / Michael Ameismeier / Jochen Baßler / Martin Beck / Roland Beckmann / Ed Hurt /  Abstract: The 5S ribonucleoprotein (RNP) is assembled from its three components (5S rRNA, Rpl5/uL18 and Rpl11/uL5) before being incorporated into the pre-60S subunit. However, when ribosome synthesis is ...The 5S ribonucleoprotein (RNP) is assembled from its three components (5S rRNA, Rpl5/uL18 and Rpl11/uL5) before being incorporated into the pre-60S subunit. However, when ribosome synthesis is disturbed, a free 5S RNP can enter the MDM2-p53 pathway to regulate cell cycle and apoptotic signaling. Here we reconstitute and determine the cryo-electron microscopy structure of the conserved hexameric 5S RNP with fungal or human factors. This reveals how the nascent 5S rRNA associates with the initial nuclear import complex Syo1-uL18-uL5 and, upon further recruitment of the nucleolar factors Rpf2 and Rrs1, develops into the 5S RNP precursor that can assemble into the pre-ribosome. In addition, we elucidate the structure of another 5S RNP intermediate, carrying the human ubiquitin ligase Mdm2, which unravels how this enzyme can be sequestered from its target substrate p53. Our data provide molecular insight into how the 5S RNP can mediate between ribosome biogenesis and cell proliferation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13134.map.gz emd_13134.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13134-v30.xml emd-13134-v30.xml emd-13134.xml emd-13134.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13134_fsc.xml emd_13134_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  emd_13134.png emd_13134.png | 68.3 KB | ||
| Filedesc metadata |  emd-13134.cif.gz emd-13134.cif.gz | 6.8 KB | ||
| Others |  emd_13134_additional_1.map.gz emd_13134_additional_1.map.gz emd_13134_additional_2.map.gz emd_13134_additional_2.map.gz emd_13134_additional_3.map.gz emd_13134_additional_3.map.gz emd_13134_additional_4.map.gz emd_13134_additional_4.map.gz | 3 MB 5 MB 21.4 MB 21.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13134 http://ftp.pdbj.org/pub/emdb/structures/EMD-13134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13134 | HTTPS FTP |
-Related structure data
| Related structure data |  7ozsMC  8bguC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13134.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13134.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hexameric 5S RNP from C. thermophilum | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: hexameric ct-sc 5S RNP monomer
| File | emd_13134_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hexameric ct-sc 5S RNP monomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
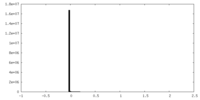
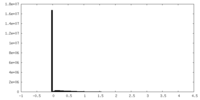
| Density Histograms |
-Additional map: hexameric ct-sc 5S RNP dimer
| File | emd_13134_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hexameric ct-sc 5S RNP dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: pre-60S with human 5S RNP
| File | emd_13134_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | pre-60S with human 5S RNP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: human MDM2-5S RNP
| File | emd_13134_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human MDM2-5S RNP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : hexameric 5S RNP from C. thermophilum
| Entire | Name: hexameric 5S RNP from C. thermophilum |
|---|---|
| Components |
|
-Supramolecule #1: hexameric 5S RNP from C. thermophilum
| Supramolecule | Name: hexameric 5S RNP from C. thermophilum / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
-Macromolecule #1: Putative ribosomal protein
| Macromolecule | Name: Putative ribosomal protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 20.119275 KDa |
| Sequence | String: MSSEKAQNPM RELRIQKLVL NISVGESGDR LTRAAKVLEQ LSGQTPVYSK ARYTVRQFGI RRNEKIAVHV TVRGPKAEEI LERGLKVKE YELRRRNFSE TGNFGFGISE HIDLGIKYDP SIGIYGMDFY CCMTRPGERV AKRRRCKSRI GASHRITREE T IRWFKQRF DGIVR UniProtKB: Large ribosomal subunit protein uL5 |
-Macromolecule #2: 60S ribosomal protein l5-like protein
| Macromolecule | Name: 60S ribosomal protein l5-like protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 35.034605 KDa |
| Sequence | String: MAFHKLVKNS AYYSRFQTKF KRRRQGKTDY YARKRLITQA KNKYNAPKYR LVVRFTNRDI ITQMVTSEIN GDKIFAAAYS HELRAYGIN HGLTNWAAAY ATGLLLARRV LAKLGLDKTF TGVEEPNGEY TLTEAAETED GERRPFKAIL DVGLARTSTG A RVFGVMKG ...String: MAFHKLVKNS AYYSRFQTKF KRRRQGKTDY YARKRLITQA KNKYNAPKYR LVVRFTNRDI ITQMVTSEIN GDKIFAAAYS HELRAYGIN HGLTNWAAAY ATGLLLARRV LAKLGLDKTF TGVEEPNGEY TLTEAAETED GERRPFKAIL DVGLARTSTG A RVFGVMKG ASDGGIFIPH SENRFPGYDI ETEELDTEVL KKYIYGGHVA EYMETLADDD EERYKSQFVK YIEDDVEADS LE ELYAEAH KQIRADPFRK YVSDAPKKSK EEWKAESLKY KKAKLSREER KARVEAKIKQ LLAEQDE UniProtKB: Large ribosomal subunit protein uL18 |
-Macromolecule #3: Ribosome biogenesis regulatory protein
| Macromolecule | Name: Ribosome biogenesis regulatory protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 22.817281 KDa |
| Sequence | String: MSTDTSKPAR LPVTVEKPTP YTFDLGLLLA NDPNPVNVPK TTDTQVLEQH LASVARDGAQ VLINQLLTTT TITATKDGVL LTLPPPTTP LPREKPVPQP KPETKWQAFA RRRGIKPKTR EQRRNLQYNP TTGQWERKWG YKGANKRGET DPIIEVSAAK E AMRPEGTS ...String: MSTDTSKPAR LPVTVEKPTP YTFDLGLLLA NDPNPVNVPK TTDTQVLEQH LASVARDGAQ VLINQLLTTT TITATKDGVL LTLPPPTTP LPREKPVPQP KPETKWQAFA RRRGIKPKTR EQRRNLQYNP TTGQWERKWG YKGANKRGET DPIIEVSAAK E AMRPEGTS VRGDKRREIR ARVKKNQKQM LRNQRIAAEK MGKK UniProtKB: Ribosome biogenesis regulatory protein homolog |
-Macromolecule #4: Ribosome production factor 2 homolog
| Macromolecule | Name: Ribosome production factor 2 homolog / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 39.92734 KDa |
| Sequence | String: MLRQIKPRNA RSKRALEKRA PKIVENPKTC LFLRGTTCSQ ITQDAMNDLY AMRQVHAKRF HKKNAIHPFE DATSLCFFSE KNDCSLMVF GSSNKKRPHT LTFVRMFDYK VLDMLEFYLD PDTYRSISQF KTSKIPIGMR PMMVFAGTAF ESPVPNAFTM A KSMLIDFF ...String: MLRQIKPRNA RSKRALEKRA PKIVENPKTC LFLRGTTCSQ ITQDAMNDLY AMRQVHAKRF HKKNAIHPFE DATSLCFFSE KNDCSLMVF GSSNKKRPHT LTFVRMFDYK VLDMLEFYLD PDTYRSISQF KTSKIPIGMR PMMVFAGTAF ESPVPNAFTM A KSMLIDFF RGEPSDKIDV EGLRFVVVVT ADEPTSSTST NNDGENPAPL PGMTDPRSID PSQKPILRLR VYGIRTKRSG TR LPRVEVE EHGPRMDFRL GRMREPDPAM LKEAMKKAKT PQEERTKKNI SMDLLGDKIG RIHMGKTDLS KLQTRKMKGL KRG RDEDEG GEDDRTDVVE AGSGEKKKKK KVKG UniProtKB: Ribosome production factor 2 homolog |
-Macromolecule #6: unknown
| Macromolecule | Name: unknown / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 1.975426 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
-Macromolecule #7: Syo1
| Macromolecule | Name: Syo1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila DSM 1495 (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 74.709156 KDa |
| Sequence | String: MGKTRRNRVR NRTDPIAKPV KPPTDPELAK LREDKILPVL KDLKSPDAKS RTTAAGAIAN IVQDAKCRKL LLREQVVHIV LTETLTDNN IDSRAAGWEI LKVLAQEEEA DFCVHLYRLD VLTAIEHAAK AVLETLTTSE PPFSKLLKAQ QRLVWDITGS L LVLIGLLA ...String: MGKTRRNRVR NRTDPIAKPV KPPTDPELAK LREDKILPVL KDLKSPDAKS RTTAAGAIAN IVQDAKCRKL LLREQVVHIV LTETLTDNN IDSRAAGWEI LKVLAQEEEA DFCVHLYRLD VLTAIEHAAK AVLETLTTSE PPFSKLLKAQ QRLVWDITGS L LVLIGLLA LARDEIHEAV ATKQTILRLL FRLISADIAP QDIYEEAISC LTTLSEDNLK VGQAITDDQE THVYDVLLKL AT GTDPRAV MACGVLHNVF TSLQWMDHSP GKDGACDAIL IPTLTRALEH VVPGGAKFNG DARYANITLL ALVTLASIGT DFQ ETLVKG NQGSRESPIS AADEEWNGFD DADGDAMDVD QKSSSGEDQE EDYEEIDVKE DDEDDDDDSI TSEMQADMER VVGA DGTDD GDLEDLPTLR ELIQTAVPQL IRLSNLPIDS DESLTIQSHA LSALNNISWT ISCLEFANGE NANIHNAWYP TAKKI WRKT ILPILEADSA DLKLATQVTS LAWAVARVLH GETPTDGNPH RKFISLYHSS KQQAGGNSNS IEEPEDPFQG LGVKCI GVV GSLAHDPAPI EVNREVGVFL VTLLRQSNNV PPAEIVEALN QLFDIYGDEE LACDKEVFWK DGFLKHLEEF LPKMRTL TK GIDKRTQPEL RTRADEALLN LGRFVQYKKK HAPK UniProtKB: Symportin 1 |
-Macromolecule #5: 5S rRNA
| Macromolecule | Name: 5S rRNA / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 38.312738 KDa |
| Sequence | String: ACGUACGACC AUACCCAGUG GAAAGCACGG CAUCCCGUCC GCUCUGCCCU AGUUAAGCCA CUGAGGGCCC GGUUAGUAGU UGGGUCGGU GACGACCAGC GAAUCCCGGG UGUUGUACGU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 43.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)