[English] 日本語
 Yorodumi
Yorodumi- EMDB-12992: SARS-CoV-2 spike subtomogram average from intracelullar viruses -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12992 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike subtomogram average from intracelullar viruses | |||||||||
 Map data Map data | SARS-CoV-2 spike subtomogram average from intracelullar viruses | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Mendonca L / Zhang P | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Authors: Luiza Mendonça / Andrew Howe / James B Gilchrist / Yuewen Sheng / Dapeng Sun / Michael L Knight / Laura C Zanetti-Domingues / Benji Bateman / Anna-Sophia Krebs / Long Chen / Julika Radecke ...Authors: Luiza Mendonça / Andrew Howe / James B Gilchrist / Yuewen Sheng / Dapeng Sun / Michael L Knight / Laura C Zanetti-Domingues / Benji Bateman / Anna-Sophia Krebs / Long Chen / Julika Radecke / Vivian D Li / Tao Ni / Ilias Kounatidis / Mohamed A Koronfel / Marta Szynkiewicz / Maria Harkiolaki / Marisa L Martin-Fernandez / William James / Peijun Zhang /   Abstract: Since the outbreak of the SARS-CoV-2 pandemic, there have been intense structural studies on purified viral components and inactivated viruses. However, structural and ultrastructural evidence on how ...Since the outbreak of the SARS-CoV-2 pandemic, there have been intense structural studies on purified viral components and inactivated viruses. However, structural and ultrastructural evidence on how the SARS-CoV-2 infection progresses in the native cellular context is scarce, and there is a lack of comprehensive knowledge on the SARS-CoV-2 replicative cycle. To correlate cytopathic events induced by SARS-CoV-2 with virus replication processes in frozen-hydrated cells, we established a unique multi-modal, multi-scale cryo-correlative platform to image SARS-CoV-2 infection in Vero cells. This platform combines serial cryoFIB/SEM volume imaging and soft X-ray cryo-tomography with cell lamellae-based cryo-electron tomography (cryoET) and subtomogram averaging. Here we report critical SARS-CoV-2 structural events - e.g. viral RNA transport portals, virus assembly intermediates, virus egress pathway, and native virus spike structures, in the context of whole-cell volumes revealing drastic cytppathic changes. This integrated approach allows a holistic view of SARS-CoV-2 infection, from the whole cell to individual molecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12992.map.gz emd_12992.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12992-v30.xml emd-12992-v30.xml emd-12992.xml emd-12992.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12992.png emd_12992.png | 51.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12992 http://ftp.pdbj.org/pub/emdb/structures/EMD-12992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12992 | HTTPS FTP |
-Validation report
| Summary document |  emd_12992_validation.pdf.gz emd_12992_validation.pdf.gz | 285.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12992_full_validation.pdf.gz emd_12992_full_validation.pdf.gz | 285.4 KB | Display | |
| Data in XML |  emd_12992_validation.xml.gz emd_12992_validation.xml.gz | 6 KB | Display | |
| Data in CIF |  emd_12992_validation.cif.gz emd_12992_validation.cif.gz | 6.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12992 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12992 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10753 (Title: Tilt-series taken from SARS-CoV-2 infected Vero cells. EMPIAR-10753 (Title: Tilt-series taken from SARS-CoV-2 infected Vero cells.Data size: 52.7 Data #1: Tilt-series taken on the periphery of SARS-CoV-2 infected Vero cells [tilt series] Data #2: Tilt-series taken on cryolamella of SARS-CoV-2 infected Vero cells [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12992.map.gz / Format: CCP4 / Size: 21.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12992.map.gz / Format: CCP4 / Size: 21.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 spike subtomogram average from intracelullar viruses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Severe acute respiratory syndrome coronavirus 2
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Severe acute respiratory syndrome coronavirus 2
| Supramolecule | Name: Severe acute respiratory syndrome coronavirus 2 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 2697049 Sci species name: Severe acute respiratory syndrome coronavirus 2 Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Details: 4% parafolmaldehyde in phosphate buffered saline |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: emClarity / Number subtomograms used: 856 |
|---|---|
| Extraction | Number tomograms: 2 / Number images used: 856 |
| CTF correction | Software - Name: emClarity (ver. 1.5.3.03) |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller


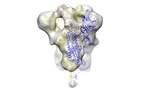











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















