+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12938 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Medicago truncatula HISN5 protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | histidine biosynthesis / herbicide design / high-resolution Cryo-EM / LYASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationimidazoleglycerol-phosphate dehydratase / imidazoleglycerol-phosphate dehydratase activity / L-histidine biosynthetic process / chloroplast Similarity search - Function | |||||||||
| Biological species |  | |||||||||
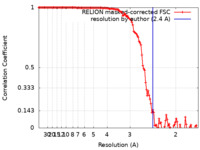
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Ruszkowski M / Witek W | |||||||||
| Funding support |  Poland, 1 items Poland, 1 items
| |||||||||
 Citation Citation |  Journal: Front Plant Sci / Year: 2024 Journal: Front Plant Sci / Year: 2024Title: Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting. Authors: Wojciech Witek / Joanna Sliwiak / Michal Rawski / Milosz Ruszkowski /  Abstract: The histidine biosynthetic pathway (HBP) is targeted for herbicide design with preliminary success only regarding imidazole-glycerol phosphate dehydratase (IGPD, EC 4.2.1.19), or HISN5, as referred ...The histidine biosynthetic pathway (HBP) is targeted for herbicide design with preliminary success only regarding imidazole-glycerol phosphate dehydratase (IGPD, EC 4.2.1.19), or HISN5, as referred to in plants. HISN5 catalyzes the sixth step of the HBP, in which imidazole-glycerol phosphate (IGP) is dehydrated to imidazole-acetol phosphate. In this work, we present high-resolution cryoEM and crystal structures of HISN5 (HISN5) in complexes with an inactive IGP diastereoisomer and with various other ligands. HISN5 can serve as a new model for plant HISN5 structural studies, as it enables resolving protein-ligand interactions at high (2.2 Å) resolution using cryoEM. We identified ligand-binding hotspots and characterized the features of plant HISN5 enzymes in the context of the HISN5-targeted inhibitor design. Virtual screening performed against millions of small molecules not only revealed candidate molecules but also identified linkers for fragments that were experimentally confirmed to bind. Based on experimental and computational approaches, this study provides guidelines for designing symmetric HISN5 inhibitors that can reach two neighboring active sites. Finally, we conducted analyses of sequence similarity networks revealing that plant HISN5 enzymes derive from cyanobacteria. We also adopted a new approach to measure HISN5 enzymatic activity using isothermal titration calorimetry and enzymatically synthesized IGP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12938.map.gz emd_12938.map.gz | 155.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12938-v30.xml emd-12938-v30.xml emd-12938.xml emd-12938.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12938_fsc.xml emd_12938_fsc.xml | 12.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12938.png emd_12938.png | 376.4 KB | ||
| Filedesc metadata |  emd-12938.cif.gz emd-12938.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12938 http://ftp.pdbj.org/pub/emdb/structures/EMD-12938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12938 | HTTPS FTP |
-Validation report
| Summary document |  emd_12938_validation.pdf.gz emd_12938_validation.pdf.gz | 446 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12938_full_validation.pdf.gz emd_12938_full_validation.pdf.gz | 445.6 KB | Display | |
| Data in XML |  emd_12938_validation.xml.gz emd_12938_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_12938_validation.cif.gz emd_12938_validation.cif.gz | 17 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12938 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12938 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12938 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12938 | HTTPS FTP |
-Related structure data
| Related structure data |  7oj5MC  8qavC  8qawC  8qaxC  8qayC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12938.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12938.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Medicago truncatula HISN5 protein
| Entire | Name: Medicago truncatula HISN5 protein |
|---|---|
| Components |
|
-Supramolecule #1: Medicago truncatula HISN5 protein
| Supramolecule | Name: Medicago truncatula HISN5 protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Imidazoleglycerol-phosphate dehydratase |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Imidazoleglycerol-phosphate dehydratase
| Macromolecule | Name: Imidazoleglycerol-phosphate dehydratase / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO / EC number: imidazoleglycerol-phosphate dehydratase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.692453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTFPIDSGA RIGEMKRVTK ETNVSVKINL DGTGVADNSS GIPFLDHMLD QLASHGLFDV HVKATGDTHI DDHHTNEDVA LAIGTALLQ ALGDRKGINR FGNFSAPLDE ALVHVSLDLS GRPHLGYDLN IPTQRVGKYD TQLVEHFFQS LVNTSGMTLH I RQFSGTNS ...String: MSTFPIDSGA RIGEMKRVTK ETNVSVKINL DGTGVADNSS GIPFLDHMLD QLASHGLFDV HVKATGDTHI DDHHTNEDVA LAIGTALLQ ALGDRKGINR FGNFSAPLDE ALVHVSLDLS GRPHLGYDLN IPTQRVGKYD TQLVEHFFQS LVNTSGMTLH I RQFSGTNS HHIIEATFKA FARALRQATE YDTRRRGTIP SSKGVLSRS UniProtKB: Imidazoleglycerol-phosphate dehydratase |
-Macromolecule #2: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 2 / Number of copies: 48 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 931 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 1008 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)