+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli 50S ribosome LiCl core particle, class 5-3 | |||||||||
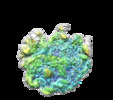
 Map data Map data | Sharpened and filtered according to the estimated local resolution | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
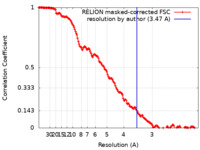
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Larsson DSD / Selmer M | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2022 Journal: Biomolecules / Year: 2022Title: Structural Consequences of Deproteinating the 50S Ribosome. Authors: Daniel S D Larsson / Sandesh Kanchugal P / Maria Selmer /  Abstract: Ribosomes are complex ribonucleoprotein particles. Purified 50S ribosomes subjected to high-salt wash, removing a subset of ribosomal proteins (r-proteins), were shown as competent for in vitro ...Ribosomes are complex ribonucleoprotein particles. Purified 50S ribosomes subjected to high-salt wash, removing a subset of ribosomal proteins (r-proteins), were shown as competent for in vitro assembly into functional 50S subunits. Here, we used cryo-EM to determine the structures of such LiCl core particles derived from 50S subunits. A wide range of complexes with large variations in the extent of the ordered 23S rRNA and the occupancy of r-proteins were resolved to between 2.8 Å and 9 Å resolution. Many of these particles showed high similarity to in vivo and in vitro assembly intermediates, supporting the inherent stability or metastability of these states. Similar to states in early ribosome assembly, the main class showed an ordered density for the particle base around the exit tunnel, with domain V and the 3'-half of domain IV disordered. In addition, smaller core particles were discovered, where either domain II or IV was unfolded. Our data support a multi-pathway in vitro disassembly process, similar but reverse to assembly. Dependencies between complex tertiary RNA structures and RNA-protein interactions were observed, where protein extensions dissociated before the globular domains. We observed the formation of a non-native RNA structure upon protein dissociation, demonstrating that r-proteins stabilize native RNA structures and prevent non-native interactions also after folding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12838.map.gz emd_12838.map.gz | 190.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12838-v30.xml emd-12838-v30.xml emd-12838.xml emd-12838.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12838_fsc.xml emd_12838_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12838.png emd_12838.png | 135 KB | ||
| Masks |  emd_12838_msk_1.map emd_12838_msk_1.map | 343 MB |  Mask map Mask map | |
| Others |  emd_12838_additional_1.map.gz emd_12838_additional_1.map.gz emd_12838_half_map_1.map.gz emd_12838_half_map_1.map.gz emd_12838_half_map_2.map.gz emd_12838_half_map_2.map.gz | 71.3 MB 276.1 MB 275.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12838 http://ftp.pdbj.org/pub/emdb/structures/EMD-12838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12838 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12838.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12838.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and filtered according to the estimated local resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_12838_msk_1.map emd_12838_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened full map, spherically masked
| File | emd_12838_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened full map, spherically masked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12838_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12838_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 50S LiCl core particle, class 1-1
| Entire | Name: 50S LiCl core particle, class 1-1 |
|---|---|
| Components |
|
-Supramolecule #1: 50S LiCl core particle, class 1-1
| Supramolecule | Name: 50S LiCl core particle, class 1-1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#16 Details: 3.5 M LiCl wash, RlmF pull-down, consensus reconstruction |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.17 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV Details: continuous carbon, 3 microL of sample, incubate for 30 seconds, blot for 3.5 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | The sample stage was tilted at 0, 15 or 30 degrees |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 11368 / Average exposure time: 0.77 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)