+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
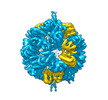
| Title | Vibrio vulnificus stressosome | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | stressosome / stress sensing / general stress response / Gram-negative / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.3 Å | ||||||||||||
 Authors Authors | Kaltwasser S / Heinz V | ||||||||||||
| Funding support | European Union,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: The Vibrio vulnificus stressosome is an oxygen-sensor involved in regulating iron metabolism Authors: Heinz V / Jackel W / Kaltwasser S / Cutugno L / Bedrunka P / Graf A / Reder A / Michalik S / Dhople VM / Madej MG / Conway M / Lechner M / Riedel K / Bange G / Boyd A / Volker U / Lewis RJ / ...Authors: Heinz V / Jackel W / Kaltwasser S / Cutugno L / Bedrunka P / Graf A / Reder A / Michalik S / Dhople VM / Madej MG / Conway M / Lechner M / Riedel K / Bange G / Boyd A / Volker U / Lewis RJ / Marles-Wright J / Ziegler C / Pane-Farre J | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12676.map.gz emd_12676.map.gz | 55.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12676-v30.xml emd-12676-v30.xml emd-12676.xml emd-12676.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12676.png emd_12676.png | 119.1 KB | ||
| Filedesc metadata |  emd-12676.cif.gz emd-12676.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12676 http://ftp.pdbj.org/pub/emdb/structures/EMD-12676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12676 | HTTPS FTP |
-Related structure data
| Related structure data |  7o0iMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12676.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12676.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.77 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : stressosome complex of VvRsbR and VvRsbS
| Entire | Name: stressosome complex of VvRsbR and VvRsbS |
|---|---|
| Components |
|
-Supramolecule #1: stressosome complex of VvRsbR and VvRsbS
| Supramolecule | Name: stressosome complex of VvRsbR and VvRsbS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
-Macromolecule #1: RsbS, negative regulator of sigma-B
| Macromolecule | Name: RsbS, negative regulator of sigma-B / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
| Molecular weight | Theoretical: 12.390511 KDa |
| Recombinant expression | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
| Sequence | String: MQSAISISKL QDVLIASVQV DLTESTLRDF SLDLLDAVKS TRAKGVMIEL SGVKTLDGES MHSLLDVVKT VEVMGRRCLL VGLRPGVVI GLMDIGIDLA STLCVADLEQ GFLYLE UniProtKB: RsbS, negative regulator of sigma-B |
-Macromolecule #2: Anti-anti-sigma factor
| Macromolecule | Name: Anti-anti-sigma factor / type: protein_or_peptide / ID: 2 / Number of copies: 48 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
| Molecular weight | Theoretical: 34.606043 KDa |
| Recombinant expression | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
| Sequence | String: MSLGSVLNKV EDADELLKLH DLTEADLALI RKFGQIMVPK LDEYVKHFYD WLRNTPEYEQ YFGDAQKLQR VQDSQVRYWK TFFDARIDS AYLKERRDVG EIHARVGLPL PTYFAGMNIS MVIFTKRMYD GSLYSDEYSS LVTAFTKLLH LDTTIVVDTY S RLINKRIS ...String: MSLGSVLNKV EDADELLKLH DLTEADLALI RKFGQIMVPK LDEYVKHFYD WLRNTPEYEQ YFGDAQKLQR VQDSQVRYWK TFFDARIDS AYLKERRDVG EIHARVGLPL PTYFAGMNIS MVIFTKRMYD GSLYSDEYSS LVTAFTKLLH LDTTIVVDTY S RLINKRIS EQSEALLAMS TPVTMIWQDI LMLPIVGIID SKRAQDIMSA VLNKISENRA KIFIMDISGV AVVDTAVANH FI KITKATK LMGCDCLVSG VSPSIARTMV QLGINVGEVR TNATLRDALE NAFKIVGLTV SGLKHFPHE UniProtKB: Anti-anti-sigma factor |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 1.5 sec. / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.5 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 59000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















