[English] 日本語
 Yorodumi
Yorodumi- EMDB-12604: Cryo-EM structure of the mycolic acid transporter MmpL3 from M. t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12604 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the mycolic acid transporter MmpL3 from M. tuberculosis | |||||||||||||||||||||
 Map data Map data | sharpened map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | TRANSPORTER / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationphosphatidylglycerol binding / Actinobacterium-type cell wall biogenesis / diacylglycerol binding / cell tip / mycolic acid biosynthetic process / cell septum / lipopolysaccharide transport / cardiolipin binding / phosphatidylethanolamine binding / membrane => GO:0016020 ...phosphatidylglycerol binding / Actinobacterium-type cell wall biogenesis / diacylglycerol binding / cell tip / mycolic acid biosynthetic process / cell septum / lipopolysaccharide transport / cardiolipin binding / phosphatidylethanolamine binding / membrane => GO:0016020 / phosphatidylinositol binding / peptidoglycan-based cell wall / regulation of membrane potential / cell wall organization / response to antibiotic / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
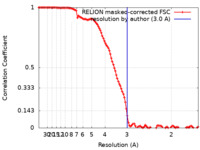
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Adams O / Deme JC | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: An emergent therapeutic target. Authors: Oliver Adams / Justin C Deme / Joanne L Parker / / Philip W Fowler / Susan M Lea / Simon Newstead /   Abstract: Tuberculosis (TB) is the leading cause of death from a single infectious agent and in 2019 an estimated 10 million people worldwide contracted the disease. Although treatments for TB exist, continual ...Tuberculosis (TB) is the leading cause of death from a single infectious agent and in 2019 an estimated 10 million people worldwide contracted the disease. Although treatments for TB exist, continual emergence of drug-resistant variants necessitates urgent development of novel antituberculars. An important new target is the lipid transporter MmpL3, which is required for construction of the unique cell envelope that shields Mycobacterium tuberculosis (Mtb) from the immune system. However, a structural understanding of the mutations in Mtb MmpL3 that confer resistance to the many preclinical leads is lacking, hampering efforts to circumvent resistance mechanisms. Here, we present the cryoelectron microscopy structure of Mtb MmpL3 and use it to comprehensively analyze the mutational landscape of drug resistance. Our data provide a rational explanation for resistance variants local to the central drug binding site, and also highlight a potential alternative route to resistance operating within the periplasmic domain. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12604.map.gz emd_12604.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12604-v30.xml emd-12604-v30.xml emd-12604.xml emd-12604.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12604_fsc.xml emd_12604_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12604.png emd_12604.png | 135.7 KB | ||
| Masks |  emd_12604_msk_1.map emd_12604_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12604.cif.gz emd-12604.cif.gz | 7.1 KB | ||
| Others |  emd_12604_additional_1.map.gz emd_12604_additional_1.map.gz emd_12604_half_map_1.map.gz emd_12604_half_map_1.map.gz emd_12604_half_map_2.map.gz emd_12604_half_map_2.map.gz | 62.6 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12604 http://ftp.pdbj.org/pub/emdb/structures/EMD-12604 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12604 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12604 | HTTPS FTP |
-Related structure data
| Related structure data |  7nvhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12604.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12604.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12604_msk_1.map emd_12604_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
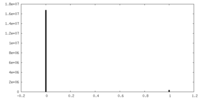
| Density Histograms |
-Additional map: unsharpened map
| File | emd_12604_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
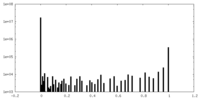
| Density Histograms |
-Half map: half map B
| File | emd_12604_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
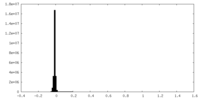
| Density Histograms |
-Half map: half map A
| File | emd_12604_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
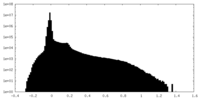
| Density Histograms |
- Sample components
Sample components
-Entire : Mycolic acid transporter MmpL3
| Entire | Name: Mycolic acid transporter MmpL3 |
|---|---|
| Components |
|
-Supramolecule #1: Mycolic acid transporter MmpL3
| Supramolecule | Name: Mycolic acid transporter MmpL3 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Trehalose monomycolate exporter MmpL3
| Macromolecule | Name: Trehalose monomycolate exporter MmpL3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.525609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFAWWGRTVY RYRFIVIGVM VALCLGGGVF GLSLGKHVTQ SGFYDDGSQS VQASVLGDQV YGRDRSGHIV AIFQAPAGKT VDDPAWSKK VVDELNRFQQ DHPDQVLGWA GYLRASQATG MATADKKYTF VSIPLKGDDD DTILNNYKAI APDLQRLDGG T VKLAGLQP ...String: MFAWWGRTVY RYRFIVIGVM VALCLGGGVF GLSLGKHVTQ SGFYDDGSQS VQASVLGDQV YGRDRSGHIV AIFQAPAGKT VDDPAWSKK VVDELNRFQQ DHPDQVLGWA GYLRASQATG MATADKKYTF VSIPLKGDDD DTILNNYKAI APDLQRLDGG T VKLAGLQP VAEALTGTIA TDQRRMEVLA LPLVAVVLFF VFGGVIAAGL PVMVGGLCIA GALGIMRFLA IFGPVHYFAQ PV VSLIGLG IAIDYGLFIV SRFREEIAEG YDTETAVRRT VITAGRTVTF SAVLIVASAI GLLLFPQGFL KSLTYATIAS VML SAILSI TVLPACLGIL GKHVDALGVR TLFRVPFLAN WKISAAYLNW LADRLQRTKT REEVEAGFWG KLVNRVMKRP VLFA APIVI IMILLIIPVG KLSLGGISEK YLPPTNSVRQ AQEEFDKLFP GYRTNPLTLV IQTSNHQPVT DAQIADIRSK AMAIG GFIE PDNDPANMWQ ERAYAVGASK DPSVRVLQNG LINPADASKK LTELRAITPP KGITVLVGGT PALELDSIHG LFAKMP LMV VILLTTTIVL MFLAFGSVVL PIKATLMSAL TLGSTMGILT WIFVDGHFSK WLNFTPTPLT APVIGLIIAL VFGLSTD YE VFLVSRMVEA RERGMSTQEA IRIGTAATGR IITAAALIVA VVAGAFVFSD LVMMKYLAFG LMAALLLDAT VVRMFLVP S VMKLLGDDCW WAPRWARRLQ TRIGLGEIHL PDGSENLYFQ UniProtKB: Trehalose monomycolate exporter MmpL3 |
-Macromolecule #2: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 2 / Number of copies: 1 / Formula: AV0 |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 58.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)