[English] 日本語
 Yorodumi
Yorodumi- EMDB-12600: Structure of the Toxoplasma gondii kinase Ron13, kinase-dead mutant -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12600 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Toxoplasma gondii kinase Ron13, kinase-dead mutant | |||||||||
 Map data Map data | Ron13 cryo-EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | secretory pathway / kinase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
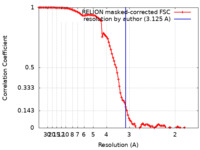
| Method | single particle reconstruction / cryo EM / Resolution: 3.125 Å | |||||||||
 Authors Authors | Korkhov VM / Mehta V | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural insights into an atypical secretory pathway kinase crucial for Toxoplasma gondii invasion. Authors: Gaëlle Lentini / Rouaa Ben Chaabene / Oscar Vadas / Chandra Ramakrishnan / Budhaditya Mukherjee / Ved Mehta / Matteo Lunghi / Jonas Grossmann / Bohumil Maco / Rémy Visentin / Adrian B Hehl ...Authors: Gaëlle Lentini / Rouaa Ben Chaabene / Oscar Vadas / Chandra Ramakrishnan / Budhaditya Mukherjee / Ved Mehta / Matteo Lunghi / Jonas Grossmann / Bohumil Maco / Rémy Visentin / Adrian B Hehl / Volodymyr M Korkhov / Dominique Soldati-Favre /   Abstract: Active host cell invasion by the obligate intracellular apicomplexan parasites relies on the formation of a moving junction, which connects parasite and host cell plasma membranes during entry. ...Active host cell invasion by the obligate intracellular apicomplexan parasites relies on the formation of a moving junction, which connects parasite and host cell plasma membranes during entry. Invading Toxoplasma gondii tachyzoites secrete their rhoptry content and insert a complex of RON proteins on the cytoplasmic side of the host cell membrane providing an anchor to which the parasite tethers. Here we show that a rhoptry-resident kinase RON13 is a key virulence factor that plays a crucial role in host cell entry. Cryo-EM, kinase assays, phosphoproteomics and cellular analyses reveal that RON13 is a secretory pathway kinase of atypical structure that phosphorylates rhoptry proteins including the components of the RON complex. Ultimately, RON13 kinase activity controls host cell invasion by anchoring the moving junction at the parasite-host cell interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12600.map.gz emd_12600.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12600-v30.xml emd-12600-v30.xml emd-12600.xml emd-12600.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12600_fsc.xml emd_12600_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12600.png emd_12600.png | 79.7 KB | ||
| Filedesc metadata |  emd-12600.cif.gz emd-12600.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12600 http://ftp.pdbj.org/pub/emdb/structures/EMD-12600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12600 | HTTPS FTP |
-Related structure data
| Related structure data |  7nurMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12600.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12600.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ron13 cryo-EM map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8544 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Secretory pathway kinase Ron13
| Entire | Name: Secretory pathway kinase Ron13 |
|---|---|
| Components |
|
-Supramolecule #1: Secretory pathway kinase Ron13
| Supramolecule | Name: Secretory pathway kinase Ron13 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein kinase domain-containing protein
| Macromolecule | Name: Protein kinase domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 126.41968 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAGTTGGKKD DQALSFLGDT KSASELNPPR LVCPDKPPTL PPREEQVRKA YALPLCELPW DDLGPMLGSG TFGRVYPLRR PACTEVTKG FVGRKFAVKI FWLKRKGMMN LFDTISQGGT PSAEQTDPGT IAAIKSEIRS LPTSSSAFRD MVRIADPTVD V EKIKGMAD ...String: GAGTTGGKKD DQALSFLGDT KSASELNPPR LVCPDKPPTL PPREEQVRKA YALPLCELPW DDLGPMLGSG TFGRVYPLRR PACTEVTKG FVGRKFAVKI FWLKRKGMMN LFDTISQGGT PSAEQTDPGT IAAIKSEIRS LPTSSSAFRD MVRIADPTVD V EKIKGMAD SLTVETIMKE AKTLRTVINT NGFYTEVGET GTIFTQMEKF VQAHRPEIWS TLSKASQEAQ ASKYAEIGLA DN HWSLPLA RVLVKDKNDV KHWALLIELF DGDLQPKTDK TGYSLDGWNA KSGGNVVLRE IFSSREALIG LTSKLVKPFV VMQ NLYSLG HFAIKPPNLL YKYFPGEKGR ASRLSVAAGD FGMAGLLHGD MILRGTLAFM APEMERVSGG LVAKPSYDVY ALAL TLASF WTAATELRDH YPWVEKCIKP TLKKMKDAPE FTFLRFASKT GPKLYEADTI YALSTCFAVG GKVEKLYHTG MPLLI RLKL SQMADPEPLA RVSMRHARFV FKAYAMLDKL LRAPQSEANA ETREEQLKQL QSLHIVQFLL FYLRMEPLTA ARDNTQ SYR RLARALLDFA RLDPVYQAAT ETVQPLPYEF FTEQKDWQNV KVEVSGSEVD ETIRKLRTSL TRDRSLSEDS WADLVDI MF GVSLDGLREV VTRVVYSRKT FLLEEKIGNA VKEAVAATYK FDPNTQLIAE DAPDRLFEVV RTDLGLSYPD DSELGRFL V HRVSKSHTAW ATVDRLARQA LRLALRREER TRQVYEQLLS GEKPSSESEK AFFDSVFSAV SVVSEANYFG LFWDFPSAG LFGVPPEEMQ AYVRKTHLAF VGKMWPVETQ KKILEAAVRV TVRGLNASLP ASLVDVYATV FAALPTKAPV SPPFLYGLER EEYSSLLFD AKLPEFKEMV AFWATRHELN IAVQTAVGKI PDATNLSDED IEKQLEGMLP AHLRSPSPAR FGWPPEAVAD N IRLFIREA KDELALHGPD MVHNRIRVNG RSKPPRRAAF LFHEIFRKAI AFKKDISVLQ FNQFFTDILK QSFDPQCRRF IA EVKKRVK SAPAEYVRVA DTEAVAPLFE GEGKDILKLV AVDPAARASD PEPNNCFLWT QAFLDDKTIV VSTGSSGSSG HHH HHHHHH H UniProtKB: Protein kinase domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 5 mM Tris pH 7.5 / 200 mM NaCl / 0.5 mM EDTA / 3 mM DTT |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)