+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacilladnavirus capsid structure | |||||||||
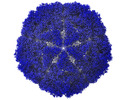
 Map data Map data | Capsid map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Jelly-roll / capsid / VIRUS | |||||||||
| Function / homology | Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Chaetoceros tenuissimus DNA virus type-II Chaetoceros tenuissimus DNA virus type-II | |||||||||
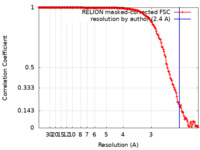
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Munke A / Okamoto K | |||||||||
| Funding support |  Sweden, Sweden,  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2022 Journal: mBio / Year: 2022Title: Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses. Authors: Anna Munke / Kei Kimura / Yuji Tomaru / Han Wang / Kazuhiro Yoshida / Seiya Mito / Yuki Hongo / Kenta Okamoto /   Abstract: Marine algae viruses are important for controlling microorganism communities in the marine ecosystem and played fundamental roles during the early events of viral evolution. Here, we have focused on ...Marine algae viruses are important for controlling microorganism communities in the marine ecosystem and played fundamental roles during the early events of viral evolution. Here, we have focused on one major group of marine algae viruses, the single-stranded DNA (ssDNA) viruses from the family. We present the capsid structure of the bacilladnavirus DNA virus type II (CtenDNAV-II), determined at 2.4-Å resolution. A structure-based phylogenetic analysis supported the previous theory that bacilladnaviruses have acquired their capsid protein via horizontal gene transfer from a ssRNA virus. The capsid protein contains the widespread virus jelly-roll fold but has additional unique features; a third β-sheet and a long C-terminal tail. Furthermore, a low-resolution reconstruction of the CtenDNAV-II genome revealed a partially spooled structure, an arrangement previously only described for dsRNA and dsDNA viruses. Together, these results exemplify the importance of genetic recombination for the emergence and evolution of ssDNA viruses and provide important insights into the underlying mechanisms that dictate genome organization. Single-stranded DNA (ssDNA) viruses are an extremely widespread group of viruses that infect diverse hosts from all three domains of life, consequently having great economic, medical, and ecological importance. In particular, bacilladnaviruses are highly abundant in marine sediments and greatly influence the dynamic appearance and disappearance of certain algae species. Despite the importance of ssDNA viruses and the last couple of years' advancements in cryo-electron microscopy, structural information on the genomes of ssDNA viruses remains limited. This paper describes two important achievements: (i) the first atomic structure of a bacilladnavirus capsid, which revealed that the capsid protein gene presumably was acquired from a ssRNA virus in early evolutionary events; and (ii) the structural organization of a ssDNA genome, which retains a spooled arrangement that previously only been observed for double-stranded viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12554.map.gz emd_12554.map.gz | 86.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12554-v30.xml emd-12554-v30.xml emd-12554.xml emd-12554.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12554_fsc.xml emd_12554_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_12554.png emd_12554.png | 296.9 KB | ||
| Masks |  emd_12554_msk_1.map emd_12554_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12554.cif.gz emd-12554.cif.gz | 5.7 KB | ||
| Others |  emd_12554_additional_1.map.gz emd_12554_additional_1.map.gz emd_12554_half_map_1.map.gz emd_12554_half_map_1.map.gz emd_12554_half_map_2.map.gz emd_12554_half_map_2.map.gz | 327.4 MB 122.4 MB 122.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12554 http://ftp.pdbj.org/pub/emdb/structures/EMD-12554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12554 | HTTPS FTP |
-Related structure data
| Related structure data |  7ns0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12554.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12554.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Capsid map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
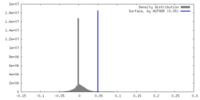
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_12554_msk_1.map emd_12554_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
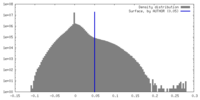
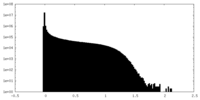
| Density Histograms |
-Additional map: Map sharpened by DeepEMhancer
| File | emd_12554_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened by DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
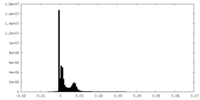
| Density Histograms |
-Half map: Capsid first half map
| File | emd_12554_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Capsid first half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
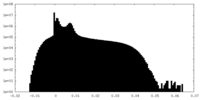
| Density Histograms |
-Half map: Capsid second half map
| File | emd_12554_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Capsid second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
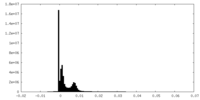
| Density Histograms |
- Sample components
Sample components
-Entire : Chaetoceros tenuissimus DNA virus type-II
| Entire | Name:  Chaetoceros tenuissimus DNA virus type-II Chaetoceros tenuissimus DNA virus type-II |
|---|---|
| Components |
|
-Supramolecule #1: Chaetoceros tenuissimus DNA virus type-II
| Supramolecule | Name: Chaetoceros tenuissimus DNA virus type-II / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Capsid / NCBI-ID: 1516127 / Sci species name: Chaetoceros tenuissimus DNA virus type-II / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Chaetoceros tenuissimus (Diatom) Chaetoceros tenuissimus (Diatom) |
| Virus shell | Shell ID: 1 / Diameter: 35.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetoceros tenuissimus DNA virus type-II Chaetoceros tenuissimus DNA virus type-II |
| Molecular weight | Theoretical: 43.329559 KDa |
| Sequence | String: MVRRRGGKTA GSKRPKMSSK NFGANRKRDF RRPARKSKAK KARSMAPAKT VRKSTTAGAH SKHFSVIGNP FSKATQQPQI PDGRMLESL PRRCQLVTEI RNNVTVGSNP TYILVAPSLG LAFQAYQDTN VPGGLDSSVY GLQNRGCTVR ANLSATSIEN Y NDIAKWRI ...String: MVRRRGGKTA GSKRPKMSSK NFGANRKRDF RRPARKSKAK KARSMAPAKT VRKSTTAGAH SKHFSVIGNP FSKATQQPQI PDGRMLESL PRRCQLVTEI RNNVTVGSNP TYILVAPSLG LAFQAYQDTN VPGGLDSSVY GLQNRGCTVR ANLSATSIEN Y NDIAKWRI VSQGINLKLN NVEDENDGWY EACRFQHDWT PDELCLRSTE NDASTISQDE DLVMGVISSS FMNGALNTIG NN MVEQRGY ESGLLKNIHK RMFQLHNNTS AIRPKTLQGQ FNYGSEITFS GTESEARFTD VPSNRQLVDS LWHNDYDCIL IKL YPRENT GAAGQTGSAL IVNAIQNLEL QYSPTSDLST YHIANKRARM VEAKLDKKNN TDAAGEPFVP GSSR UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)