+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the autoinducer-2 exporter TqsA from E. coli | |||||||||
 Map data Map data | C5 symmetry imposed final map for TqsA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-EM / membrane protein / autoinducer-2 exporter / quorum sensing / pentamer / protein oligomerization / STRUCTURAL PROTEIN | |||||||||
| Function / homology | autoinducer AI-2 transmembrane transport / Transmembrane protein TqsA-like / AI-2E family transporter / quorum sensing / efflux transmembrane transporter activity / transmembrane transport / plasma membrane / AI-2 transport protein TqsA Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
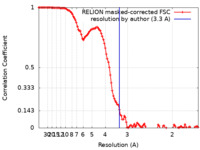
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Khera R / Xie H | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2022 Journal: EMBO J / Year: 2022Title: Cryo-EM structures of pentameric autoinducer-2 exporter from Escherichia coli reveal its transport mechanism. Authors: Radhika Khera / Ahmad R Mehdipour / Jani R Bolla / Joerg Kahnt / Sonja Welsch / Ulrich Ermler / Cornelia Muenke / Carol V Robinson / Gerhard Hummer / Hao Xie / Hartmut Michel /    Abstract: Bacteria utilize small extracellular molecules to communicate in order to collectively coordinate their behaviors in response to the population density. Autoinducer-2 (AI-2), a universal molecule for ...Bacteria utilize small extracellular molecules to communicate in order to collectively coordinate their behaviors in response to the population density. Autoinducer-2 (AI-2), a universal molecule for both intra- and inter-species communication, is involved in the regulation of biofilm formation, virulence, motility, chemotaxis, and antibiotic resistance. While many studies have been devoted to understanding the biosynthesis and sensing of AI-2, very little information is available on its export. The protein TqsA from Escherichia coli, which belongs to the AI-2 exporter superfamily, has been shown to export AI-2. Here, we report the cryogenic electron microscopic structures of two AI-2 exporters (TqsA and YdiK) from E. coli at 3.35 Å and 2.80 Å resolutions, respectively. Our structures suggest that the AI-2 exporter exists as a homo-pentameric complex. In silico molecular docking and native mass spectrometry experiments were employed to demonstrate the interaction between AI-2 and TqsA, and the results highlight the functional importance of two helical hairpins in substrate binding. We propose that each monomer works as an independent functional unit utilizing an elevator-type transport mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12256.map.gz emd_12256.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12256-v30.xml emd-12256-v30.xml emd-12256.xml emd-12256.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12256_fsc.xml emd_12256_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12256.png emd_12256.png | 107 KB | ||
| Masks |  emd_12256_msk_1.map emd_12256_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12256.cif.gz emd-12256.cif.gz | 5.9 KB | ||
| Others |  emd_12256_additional_1.map.gz emd_12256_additional_1.map.gz emd_12256_half_map_1.map.gz emd_12256_half_map_1.map.gz emd_12256_half_map_2.map.gz emd_12256_half_map_2.map.gz | 13.5 MB 98.3 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12256 http://ftp.pdbj.org/pub/emdb/structures/EMD-12256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12256 | HTTPS FTP |
-Related structure data
| Related structure data |  7nb6MC  7ot9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12256.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12256.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C5 symmetry imposed final map for TqsA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
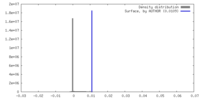
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_12256_msk_1.map emd_12256_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
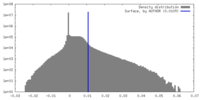
| Density Histograms |
-Additional map: C1 symmetry map
| File | emd_12256_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C1 symmetry map | ||||||||||||
| Projections & Slices |
| ||||||||||||
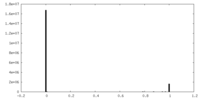
| Density Histograms |
-Half map: Half map2
| File | emd_12256_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
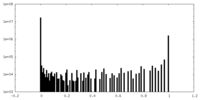
| Density Histograms |
-Half map: Half map1
| File | emd_12256_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pentameric TqsA
| Entire | Name: Pentameric TqsA |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric TqsA
| Supramolecule | Name: Pentameric TqsA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 187 KDa |
-Macromolecule #1: AI-2 transport protein TqsA
| Macromolecule | Name: AI-2 transport protein TqsA / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.563543 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKPIITLNG LKIVIMLGML VIILCGIRFA AEIIVPFILA LFIAVILNPL VQHMVRWRVP RVLAVSILMT IIVMAMVLLL AYLGSALNE LTRTLPQYRN SIMTPLQALE PLLQRVGIDV SVDQLAHYID PNAAMTLLTN LLTQLSNAMS SIFLLLLTVL F MLLEVPQL ...String: MAKPIITLNG LKIVIMLGML VIILCGIRFA AEIIVPFILA LFIAVILNPL VQHMVRWRVP RVLAVSILMT IIVMAMVLLL AYLGSALNE LTRTLPQYRN SIMTPLQALE PLLQRVGIDV SVDQLAHYID PNAAMTLLTN LLTQLSNAMS SIFLLLLTVL F MLLEVPQL PGKFQQMMAR PVEGMAAIQR AIDSVSHYLV LKTAISIITG LVAWAMLAAL DVRFAFVWGL LAFALNYIPN IG SVLAAIP PIAQVLVFNG FYEALLVLAG YLLINLVFGN ILEPRIMGRG LGLSTLVVFL SLIFWGWLLG PVGMLLSVPL TII VKIALE QTAGGQSIAV LLSDLNKE UniProtKB: AI-2 transport protein TqsA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50 mM Tris (pH 7.5), 150 mM NaCl and 0.006% GDN |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK III / Details: blot time 4 s, blot force +20. |
| Details | Its a membrane protein and for purification, glyco-diosgenin was used. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 5452 / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)