[English] 日本語
 Yorodumi
Yorodumi- EMDB-1222: Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals or... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1222 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. | |||||||||
 Map data Map data | This is the map of bacteriophage P22. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Chang J / Weigele P / King J / Chiu W / Jiang W | |||||||||
 Citation Citation |  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006Title: Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Authors: Juan Chang / Peter Weigele / Jonathan King / Wah Chiu / Wen Jiang /  Abstract: The mechanisms by which most double-stranded DNA viruses package and release their genomic DNA are not fully understood. Single particle cryo-electron microscopy and asymmetric 3D reconstruction ...The mechanisms by which most double-stranded DNA viruses package and release their genomic DNA are not fully understood. Single particle cryo-electron microscopy and asymmetric 3D reconstruction reveal the organization of the complete bacteriophage P22 virion, including the protein channel through which DNA is first packaged and later ejected. This channel is formed by a dodecamer of portal proteins and sealed by a tail hub consisting of two stacked barrels capped by a protein needle. Six trimeric tailspikes attached around this tail hub are kinked, suggesting a functional hinge that may be used to trigger DNA release. Inside the capsid, the portal's central channel is plugged by densities interpreted as pilot/injection proteins. A short rod-like density near these proteins may be the terminal segment of the dsDNA genome. The coaxially packed DNA genome is encapsidated by the icosahedral shell. This complete structure unifies various biochemical, genetic, and crystallographic data of its components from the past several decades. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1222.map.gz emd_1222.map.gz | 84.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1222-v30.xml emd-1222-v30.xml emd-1222.xml emd-1222.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  1222.gif 1222.gif | 34.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1222 http://ftp.pdbj.org/pub/emdb/structures/EMD-1222 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1222 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1222 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1222.map.gz / Format: CCP4 / Size: 89 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1222.map.gz / Format: CCP4 / Size: 89 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the map of bacteriophage P22. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
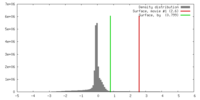
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage P22
| Entire | Name:  Bacteriophage P22 (virus) Bacteriophage P22 (virus) |
|---|---|
| Components |
|
-Supramolecule #1000: Bacteriophage P22
| Supramolecule | Name: Bacteriophage P22 / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Enterobacteria phage P22
| Supramolecule | Name: Enterobacteria phage P22 / type: virus / ID: 1 / Name.synonym: P22 / NCBI-ID: 10754 / Sci species name: Enterobacteria phage P22 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: P22 |
|---|---|
| Host (natural) | Organism:  Salmonella (bacteria) / synonym: BACTERIA(EUBACTERIA) Salmonella (bacteria) / synonym: BACTERIA(EUBACTERIA) |
| Virus shell | Shell ID: 1 / Name: gp5 / Diameter: 700 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 10 mM Tris pH 7.5, 1mM MgCl2 |
|---|---|
| Grid | Details: 200 mesh copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Temperature | Average: 100 K |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Side entry / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 16000 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)