[English] 日本語
 Yorodumi
Yorodumi- EMDB-11965: A reconstruction of Puumala virus-like particle glycoprotein spik... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11965 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A reconstruction of Puumala virus-like particle glycoprotein spike lattice in the presence of fab 4G2 which is absent from the reconstruction. | |||||||||
 Map data Map data | A reconstruction of Puumala virus-like particle glycoprotein spike lattice in the presence of fab 4G2 which is absent from the reconstruction. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Puumala virus - Sotkamo Puumala virus - Sotkamo | |||||||||
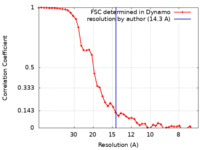
| Method | subtomogram averaging / cryo EM / Resolution: 14.3 Å | |||||||||
 Authors Authors | Rissanen I / Stass R / Huiskonen JT / Bowden TA | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein. Authors: Ilona Rissanen / Robert Stass / Stefanie A Krumm / Jeffrey Seow / Ruben Jg Hulswit / Guido C Paesen / Jussi Hepojoki / Olli Vapalahti / Åke Lundkvist / Olivier Reynard / Viktor Volchkov / ...Authors: Ilona Rissanen / Robert Stass / Stefanie A Krumm / Jeffrey Seow / Ruben Jg Hulswit / Guido C Paesen / Jussi Hepojoki / Olli Vapalahti / Åke Lundkvist / Olivier Reynard / Viktor Volchkov / Katie J Doores / Juha T Huiskonen / Thomas A Bowden /      Abstract: The intricate lattice of Gn and Gc glycoprotein spike complexes on the hantavirus envelope facilitates host-cell entry and is the primary target of the neutralizing antibody-mediated immune response. ...The intricate lattice of Gn and Gc glycoprotein spike complexes on the hantavirus envelope facilitates host-cell entry and is the primary target of the neutralizing antibody-mediated immune response. Through study of a neutralizing monoclonal antibody termed mAb P-4G2, which neutralizes the zoonotic pathogen Puumala virus (PUUV), we provide a molecular-level basis for antibody-mediated targeting of the hantaviral glycoprotein lattice. Crystallographic analysis demonstrates that P-4G2 binds to a multi-domain site on PUUV Gc and may preclude fusogenic rearrangements of the glycoprotein that are required for host-cell entry. Furthermore, cryo-electron microscopy of PUUV-like particles in the presence of P-4G2 reveals a lattice-independent configuration of the Gc, demonstrating that P-4G2 perturbs the (Gn-Gc) lattice. This work provides a structure-based blueprint for rationalizing antibody-mediated targeting of hantaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11965.map.gz emd_11965.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11965-v30.xml emd-11965-v30.xml emd-11965.xml emd-11965.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11965_fsc.xml emd_11965_fsc.xml | 4.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11965.png emd_11965.png | 111.9 KB | ||
| Masks |  emd_11965_msk_1.map emd_11965_msk_1.map | 6.9 MB |  Mask map Mask map | |
| Others |  emd_11965_half_map_1.map.gz emd_11965_half_map_1.map.gz emd_11965_half_map_2.map.gz emd_11965_half_map_2.map.gz | 5.7 MB 5.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11965 http://ftp.pdbj.org/pub/emdb/structures/EMD-11965 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11965 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11965 | HTTPS FTP |
-Validation report
| Summary document |  emd_11965_validation.pdf.gz emd_11965_validation.pdf.gz | 535.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11965_full_validation.pdf.gz emd_11965_full_validation.pdf.gz | 534.4 KB | Display | |
| Data in XML |  emd_11965_validation.xml.gz emd_11965_validation.xml.gz | 10.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11965 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11965 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11965 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11965 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11965.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11965.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A reconstruction of Puumala virus-like particle glycoprotein spike lattice in the presence of fab 4G2 which is absent from the reconstruction. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11965_msk_1.map emd_11965_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
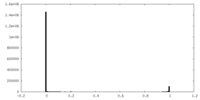
| Density Histograms |
-Half map: Half map (even) used to estimate resolution by FSC.
| File | emd_11965_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (even) used to estimate resolution by FSC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
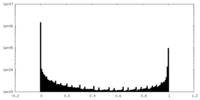
| Density Histograms |
-Half map: Half map (odd) used to estimate resolution by FSC.
| File | emd_11965_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (odd) used to estimate resolution by FSC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
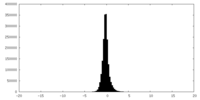
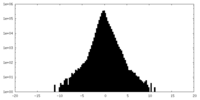
| Density Histograms |
- Sample components
Sample components
-Entire : Puumala virus - Sotkamo
| Entire | Name:  Puumala virus - Sotkamo Puumala virus - Sotkamo |
|---|---|
| Components |
|
-Supramolecule #1: Puumala virus - Sotkamo
| Supramolecule | Name: Puumala virus - Sotkamo / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 11606 / Sci species name: Puumala virus - Sotkamo / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Myodes glareolus (Bank vole) Myodes glareolus (Bank vole) |
| Host system | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: PBS |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 4.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)