[English] 日本語
 Yorodumi
Yorodumi- EMDB-11905: In situ subtomogram averaging structure of the type III secretion... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11905 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ subtomogram averaging structure of the type III secretion system of yersinia enterocolitica - sorting platform | |||||||||
 Map data Map data | Subtomogram average structure of the sorting platform of the type III secretion system of yersinia enterocolitica with C6 symmetry applied, filted to 4 nm resolution. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 40.0 Å | |||||||||
 Authors Authors | Berger C / Ravelli RBG / Lopez-Iglesias C / Kudryashev M / Diepold A / Peters PJ | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2021 Journal: J Struct Biol / Year: 2021Title: Structure of the Yersinia injectisome in intracellular host cell phagosomes revealed by cryo FIB electron tomography. Authors: Casper Berger / Raimond B G Ravelli / Carmen López-Iglesias / Mikhail Kudryashev / Andreas Diepold / Peter J Peters /   Abstract: Many pathogenic bacteria use the type III secretion system (T3SS), or injectisome, to secrete toxins into host cells. These protruding systems are primary targets for drug and vaccine development. ...Many pathogenic bacteria use the type III secretion system (T3SS), or injectisome, to secrete toxins into host cells. These protruding systems are primary targets for drug and vaccine development. Upon contact between injectisomes and host membranes, toxin secretion is triggered. How this works structurally and functionally is yet unknown. Using cryo-focused ion beam milling and cryo-electron tomography, we visualized injectisomes of Yersinia enterocolitica inside the phagosomes of infected human myeloid cells in a close-to-native state. We observed that a minimum needle length is required for injectisomes to contact the host membrane and bending of host membranes by some injectisomes that contact the host. Through subtomogram averaging, the structure of the entire injectisome was determined, which revealed structural differences in the cytosolic sorting platform compared to other bacteria. These findings contribute to understanding how injectisomes secrete toxins into host cells and provides the indispensable native context. The application of these cryo-electron microscopy techniques paves the way for the study of the 3D structure of infection-relevant protein complexes in host-pathogen interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11905.map.gz emd_11905.map.gz | 58.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11905-v30.xml emd-11905-v30.xml emd-11905.xml emd-11905.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11905_fsc.xml emd_11905_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11905.png emd_11905.png | 77.8 KB | ||
| Masks |  emd_11905_msk_1.map emd_11905_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_11905_additional_1.map.gz emd_11905_additional_1.map.gz emd_11905_half_map_1.map.gz emd_11905_half_map_1.map.gz emd_11905_half_map_2.map.gz emd_11905_half_map_2.map.gz | 58.4 MB 58.4 MB 58.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11905 http://ftp.pdbj.org/pub/emdb/structures/EMD-11905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11905 | HTTPS FTP |
-Validation report
| Summary document |  emd_11905_validation.pdf.gz emd_11905_validation.pdf.gz | 826.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11905_full_validation.pdf.gz emd_11905_full_validation.pdf.gz | 825.5 KB | Display | |
| Data in XML |  emd_11905_validation.xml.gz emd_11905_validation.xml.gz | 14.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11905 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11905 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11905 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11905 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11905.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11905.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average structure of the sorting platform of the type III secretion system of yersinia enterocolitica with C6 symmetry applied, filted to 4 nm resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.452 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
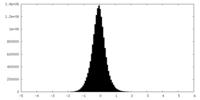
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11905_msk_1.map emd_11905_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
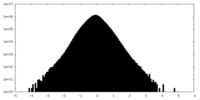
| Density Histograms |
-Additional map: Subtomogram average structure of the sorting platform of...
| File | emd_11905_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average structure of the sorting platform of the type III secretion system of yersinia enterocolitica with C6 symmetry applied. | ||||||||||||
| Projections & Slices |
| ||||||||||||
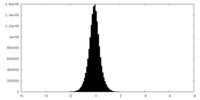
| Density Histograms |
-Half map: half-map used to determine the resolution with C6 symmetry applied
| File | emd_11905_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map used to determine the resolution with C6 symmetry applied | ||||||||||||
| Projections & Slices |
| ||||||||||||
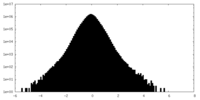
| Density Histograms |
-Half map: half-map used to determine the resolution with C6 symmetry applied
| File | emd_11905_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map used to determine the resolution with C6 symmetry applied | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sorting platform of the Type III secretion system of Yersinia ent...
| Entire | Name: Sorting platform of the Type III secretion system of Yersinia enterocolitica |
|---|---|
| Components |
|
-Supramolecule #1: Sorting platform of the Type III secretion system of Yersinia ent...
| Supramolecule | Name: Sorting platform of the Type III secretion system of Yersinia enterocolitica type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) / Strain: E40 Yersinia enterocolitica (bacteria) / Strain: E40 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: CElls were grown in RPMI 1640 medium on the grid. 2 ul of RPMI 1640 was applied to the grid just prior to vitrification |
|---|---|
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 310 K / Instrument: HOMEMADE PLUNGER Details: FEI vitrobot modified with a jet-vitrification module and a blotforce feedback mechanism.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 24.0 sec. / Average electron dose: 2.86 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: -0.005 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)