+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11859 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | yeast THO-Sub2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | yeast THO complex S. cerevisiae THO-Sub2 the transcription-export (TREX) complex / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleoplasmic THO complex / cellular response to azide / THO complex / THO complex part of transcription export complex / positive regulation of transcription elongation by RNA polymerase I / transcription export complex / Cdc73/Paf1 complex / mRNA 3'-end processing / positive regulation of transcription by RNA polymerase I / subtelomeric heterochromatin formation ...nucleoplasmic THO complex / cellular response to azide / THO complex / THO complex part of transcription export complex / positive regulation of transcription elongation by RNA polymerase I / transcription export complex / Cdc73/Paf1 complex / mRNA 3'-end processing / positive regulation of transcription by RNA polymerase I / subtelomeric heterochromatin formation / mRNA export from nucleus / transcription-coupled nucleotide-excision repair / stress granule assembly / spliceosomal complex / transcription elongation by RNA polymerase II / mRNA splicing, via spliceosome / mRNA processing / DNA recombination / nucleic acid binding / molecular adaptor activity / chromosome, telomeric region / RNA helicase activity / RNA helicase / mRNA binding / ATP hydrolysis activity / RNA binding / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Schuller SK / Schuller JM | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex. Authors: Sandra K Schuller / Jan M Schuller / J Rajan Prabu / Marc Baumgärtner / Fabien Bonneau / Jérôme Basquin / Elena Conti /  Abstract: The yeast THO complex is recruited to active genes and interacts with the RNA-dependent ATPase Sub2 to facilitate the formation of mature export-competent messenger ribonucleoprotein particles and to ...The yeast THO complex is recruited to active genes and interacts with the RNA-dependent ATPase Sub2 to facilitate the formation of mature export-competent messenger ribonucleoprotein particles and to prevent the co-transcriptional formation of RNA:DNA-hybrid-containing structures. How THO-containing complexes function at the mechanistic level is unclear. Here, we elucidated a 3.4 Å resolution structure of THO-Sub2 by cryo-electron microscopy. THO subunits Tho2 and Hpr1 intertwine to form a platform that is bound by Mft1, Thp2, and Tex1. The resulting complex homodimerizes in an asymmetric fashion, with a Sub2 molecule attached to each protomer. The homodimerization interfaces serve as a fulcrum for a seesaw-like movement concomitant with conformational changes of the Sub2 ATPase. The overall structural architecture and topology suggest the molecular mechanisms of nucleic acid remodeling during mRNA biogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11859.map.gz emd_11859.map.gz | 65.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11859-v30.xml emd-11859-v30.xml emd-11859.xml emd-11859.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11859.png emd_11859.png | 153.5 KB | ||
| Filedesc metadata |  emd-11859.cif.gz emd-11859.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11859 http://ftp.pdbj.org/pub/emdb/structures/EMD-11859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11859 | HTTPS FTP |
-Related structure data
| Related structure data |  7apxMC  7aqoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11859.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11859.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : yeast THO-Sub2 complex
| Entire | Name: yeast THO-Sub2 complex |
|---|---|
| Components |
|
-Supramolecule #1: yeast THO-Sub2 complex
| Supramolecule | Name: yeast THO-Sub2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 431 kDa/nm |
-Macromolecule #1: THO complex subunit 2,Tho2
| Macromolecule | Name: THO complex subunit 2,Tho2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 186.067094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAASMAEQTL LSKLNALSQK VIPPASPSQA SILTEEVIRN WPERSKTLCS DFTALESNDE KEDWLRTLFI ELFDFINKND ENSPLKLSD VASFTNELVN HERQVSQASI VGKMFIAVSS TVPNINDLTT ISLCKLIPSL HEELFKFSWI SSKLLNKEQT T LLRHLLKK ...String: GAASMAEQTL LSKLNALSQK VIPPASPSQA SILTEEVIRN WPERSKTLCS DFTALESNDE KEDWLRTLFI ELFDFINKND ENSPLKLSD VASFTNELVN HERQVSQASI VGKMFIAVSS TVPNINDLTT ISLCKLIPSL HEELFKFSWI SSKLLNKEQT T LLRHLLKK SKYELKKYNL LVENSVGYGQ LVALLILAYY DPDNFSKVSA YLKEIYHIMG KYSLDSIRTL DVILNVSSQF IT EGYKFFI ALLRKSDSWP SSHVANNSNY SSLNEGGNMI AANIISFNLS QYNEEVDKEN YERYMDMCCI LLKNGFVNFY SIW DNVKPE MEFLQEYIQN LETELEEEST KGVENPLAMA AALSTENETD EDNALVVNDD VNMKDKISEE TNADIESKGK QKTQ QDILL FGKIKLLERL LIHGCVIPVI HVLKQYPKVL YVSESLSRYL GRVFEYLLNP LYTSMTSSGE SKDMATALMI TRIDN GILA HKPRLIHKYK THEPFESLEL NSSYVFYYSE WNSNLTPFAS VNDLFENSHI YLSIIGPYLG RIPTLLSKIS RIGVAD IQK NHGSESLHVT IDKWIDYVRK FIFPATSLLQ NNPIATSEVY ELMKFFPFEK RYFIYNEMMT KLSQDILPLK VSFNKAE RE AKSILKALSI DTIAKESRRF AKLISTNPLA SLVPAVKQIE NYDKVSELVV YTTKYFNDFA YDVLQFVLLL RLTYNRPA V QFDGVNQAMW VQRLSIFIAG LAKNCPNMDI SNIITYILKT LHNGNIIAVS ILKELIITVG GIRDLNEVNM KQLLMLNSG SPLKQYARHL IYDFRDDNSV ISSRLTSFFT DQSAISEIIL LLYTLNLKAN TQNSHYKILS TRCDEMNTLL WSFIELIKHC LKGKAFEEN VLPFVELNNR FHLSTPWTFH IWRDYLDNQL NSNENFSIDE LIEGAEFSDV DLTKISKDLF TTFWRLSLYD I HFDKSLYD ERKNALSGEN TGHMSNRKKH LIQNQIKDIL VTGISHQRAF KKTSEFISEK SNVWNKDCGE DQIKIFLQNC VV PRVLFSP SDALFSSFFI FMAFRTENLM SILNTCITSN ILKTLLFCCT SSEAGNLGLF FTDVLKKLEK MRLNGDFNDQ ASR KLYEWH SVITEQVIDL LSEKNYMSIR NGIEFMKHVT SVFPVVKAHI QLVYTTLEEN LINEEREDIK LPSSALIGHL KARL KDALE LDEFCTLTEE EAEQKRIREM ELEEIKNYET ACQNEQKQVA LRKQLELNKS QRLQNDPPKS VASGSAGLNS KDRYT YSRN EPVIPTKPSS SQWSYSKVTR HVDDINHYLA TNHLQKAISL VENDDETRNL RKLSKQNMPI FDFRNSTLEI FERYFR TLI QNPQNPDFAE KIDSLKRYIK NISREPYPDT TSSYSEAAAP EYTKRSSRYS GNAGGKDGYG SSNYRGPSND RSAPKNI KP ISSYAHKRSE LPTRPSKSKT YNDRSRALRP TGPDRGDGFD QRDNRLREEY KKNSSQRSQL RFPEKPFQEG KDSSKANP Y QASSYKRDSP SENEEKPNKR FKKDETIRNK FQTQDYRNTR DSGAAHRANE NQRYNGNRKS NTQALPQGPK GGNYVSRYQ R(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK) UniProtKB: THO complex subunit 2 |
-Macromolecule #2: THO complex subunit HPR1
| Macromolecule | Name: THO complex subunit HPR1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 84.544047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNTEELIQN SIGFLQKTFK ALPVSFDSIR HEPLPSSMLH ASVLNFEWEP LEKNISAIHD RDSLIDIILK RFIIDSMTNA IEDEEENNL EKGLLNSCIG LDFVYNSRFN RSNPASWGNT FFELFSTIID LLNSPSTFLK FWPYAESRIE WFKMNTSVEP V SLGESNLI ...String: MSNTEELIQN SIGFLQKTFK ALPVSFDSIR HEPLPSSMLH ASVLNFEWEP LEKNISAIHD RDSLIDIILK RFIIDSMTNA IEDEEENNL EKGLLNSCIG LDFVYNSRFN RSNPASWGNT FFELFSTIID LLNSPSTFLK FWPYAESRIE WFKMNTSVEP V SLGESNLI SYKQPLYEKL RHWNDILAKL ENNDILNTVK HYNMKYKLEN FLSELLPINE ESNFNRSASI SALQESDNEW NR SARERES NRSSDVIFAA DYNFVFYHLI ICPIEFAFSD LEYKNDVDRS LSPLLDAILE IEENFYSKIK MNNRTRYSLE EAL NTEYYA NYDVMTPKLP VYMKHSNAMK MDRNEFWANL QNIKESDDYT LRPTIMDISL SNTTCLYKQL TQEDDDYYRK QFIL QLCFT TNLIRNLISS DETRNFYKSC YLRENPLSDI DFENLDEVNK KRGLNLCSYI CDNRVLKFYK IKDPDFYRVI RKLMS SDEK FTTAKIDGFK EFQNFRISKE KIPPPAFDET FKKFTFIKMG NKLINNVWKI PTGLDKIEQE VKKPEGVYEA AQAKWE SKI SSETSGGEAK DEIIRQWQTL RFLRSRYLFD FDKVNEKTGV DGLFEEPRKV EALDDSFKEK LLYKINQEHR KKLQDAR EY KIGKERKKRA LEEEASFPER EQKIKSQRIN SASQTEGDEL KSEQTQPKGE ISEENTKIKS SEVSSQDPDS GVAGEFAP UniProtKB: THO complex subunit HPR1 |
-Macromolecule #3: THO complex subunit THP2
| Macromolecule | Name: THO complex subunit THP2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 30.340264 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKEEGRTYF ESLCEEEQSL QESQTHLLNI LDILSVLADP RSSDDLLTES LKKLPDLHRE LINSSIRLRY DKYQTREAQL LEDTKTGRD VAAGVQNPKS ISEYYSTFEH LNRDTLRYIN LLKRLSVDLA KQVEVSDPSV TVYEMDKWVP SEKLQGILEQ Y CAPDTDIR ...String: MTKEEGRTYF ESLCEEEQSL QESQTHLLNI LDILSVLADP RSSDDLLTES LKKLPDLHRE LINSSIRLRY DKYQTREAQL LEDTKTGRD VAAGVQNPKS ISEYYSTFEH LNRDTLRYIN LLKRLSVDLA KQVEVSDPSV TVYEMDKWVP SEKLQGILEQ Y CAPDTDIR GVDAQIKNYL DQIKMARAKF GLENKYSLKE RLSTLTKELN HWRKEWDDIE MLMFGDDAHS MKKMIQKIDS LK SEINAPS ESYPVDKEGD IVLE UniProtKB: THO complex subunit THP2 |
-Macromolecule #4: THO complex subunit MFT1
| Macromolecule | Name: THO complex subunit MFT1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 45.055012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPLSQKQIDQ VRTKVHYSEV DTPFNKYLDI LGKVTKLTGS IINGTLSNDD SKIEKLTEQN ISQLKESAHL RFLDLQSSID TKKVADENW ETCQQETLAK LENLKDKLPD IKSIHSKLLL RIGKLQGLYD SVQVINREVE GLSEGRTSLV VTRAEWEKEL G TDLVKFLI ...String: MPLSQKQIDQ VRTKVHYSEV DTPFNKYLDI LGKVTKLTGS IINGTLSNDD SKIEKLTEQN ISQLKESAHL RFLDLQSSID TKKVADENW ETCQQETLAK LENLKDKLPD IKSIHSKLLL RIGKLQGLYD SVQVINREVE GLSEGRTSLV VTRAEWEKEL G TDLVKFLI EKNYLKLVDP GLKKDSSEER YRIYDDFSKG PKELESINAS MKSDIENVRQ EVSSYKEKWL RDAEIFGKIT SI FKEELLK RDGLLNEAEG DNIDEDYESD EDEERKERFK RQRSMVEVNT IENVDEKEES DHEYDDQEDE ENEEEDDMEV DVE DIKEDN EVDGESSQQE DNSRQGNNEE TDKETGVIEE PDAVNDAEEA DSDHSSRKLG GTTSDFSASS SVEEVK UniProtKB: THO complex subunit MFT1 |
-Macromolecule #5: Protein TEX1
| Macromolecule | Name: Protein TEX1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 42.315168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTIGAVDIL NQKTITSEVA ASVTSKYLQS TFSKGNTSHI EDKRFIHVSS RSHSRFTSTP ITPNEILSLK FHVSGSSMAY SRMDGSLTV WFIKDASFDK SVEVYIPDCC GSDKLATDLS WNPTSLNQIA VVSNSSEISL LLINEKSLTA SKLRTLSLGS K TKVNTCLY ...String: MSTIGAVDIL NQKTITSEVA ASVTSKYLQS TFSKGNTSHI EDKRFIHVSS RSHSRFTSTP ITPNEILSLK FHVSGSSMAY SRMDGSLTV WFIKDASFDK SVEVYIPDCC GSDKLATDLS WNPTSLNQIA VVSNSSEISL LLINEKSLTA SKLRTLSLGS K TKVNTCLY DPLGNWLLAA TKSEKIYLFD VKKDHSSVCS LNISDISQED NDVVYSLAWS NGGSHIFIGF KSGYLAILKA KH GILEVCT KIKAHTGPIT EIKMDPWGRN FITGSIDGNC YVWNMKSLCC ELIINDLNSA VTTLDVCHLG KILGICTEDE MVY FYDLNS GNLLHSKSLA NYKTDPVLKF YPDKSWYIMS GKNDTLSNHF VKNEKNLITY WKDM UniProtKB: Protein TEX1 |
-Macromolecule #6: ATP-dependent RNA helicase SUB2
| Macromolecule | Name: ATP-dependent RNA helicase SUB2 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 45.521004 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAASDKKGSY VGIHSTGFKD FLLKPELSRA IIDCGFEHPS EVQQHTIPQS IHGTDVLCQA KSGLGKTAVF VLSTLQQLDP VPGEVAVVV ICNARELAYQ IRNEYLRFSK YMPDVKTAVF YGGTPISKDA ELLKNKDTAP HIVVATPGRL KALVREKYID L SHVKNFVI ...String: GAASDKKGSY VGIHSTGFKD FLLKPELSRA IIDCGFEHPS EVQQHTIPQS IHGTDVLCQA KSGLGKTAVF VLSTLQQLDP VPGEVAVVV ICNARELAYQ IRNEYLRFSK YMPDVKTAVF YGGTPISKDA ELLKNKDTAP HIVVATPGRL KALVREKYID L SHVKNFVI DECDKVLEEL DMRRDVQEIF RATPRDKQVM MFSATLSQEI RPICRRFLQN PLEIFVDDEA KLTLHGLQQY YI KLEEREK NRKLAQLLDD LEFNQVIIFV KSTTRANELT KLLNASNFPA ITVHGHMKQE ERIARYKAFK DFEKRICVST DVF GRGIDI ERINLAINYD LTNEADQYLH RVGRAGRFGT KGLAISFVSS KEDEEVLAKI QERFDVKIAE FPEEGIDPST YLNN UniProtKB: ATP-dependent RNA helicase SUB2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7apx: |
 Movie
Movie Controller
Controller



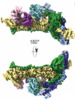









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















