+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11689 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T275P after heme uptake from M. tuberculosis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Heme / Catalase / Peroxidase enzyme / METAL BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxidoreductase activity, acting on a heme group of donors, nitrogenous group as acceptor / Tolerance of reactive oxygen produced by macrophages / catalase-peroxidase / NADH binding / catalase activity / NADPH binding / peptidoglycan-based cell wall / positive regulation of DNA repair / hydrogen peroxide catabolic process / peroxidase activity ...oxidoreductase activity, acting on a heme group of donors, nitrogenous group as acceptor / Tolerance of reactive oxygen produced by macrophages / catalase-peroxidase / NADH binding / catalase activity / NADPH binding / peptidoglycan-based cell wall / positive regulation of DNA repair / hydrogen peroxide catabolic process / peroxidase activity / cellular response to hydrogen peroxide / response to oxidative stress / response to antibiotic / heme binding / extracellular region / metal ion binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
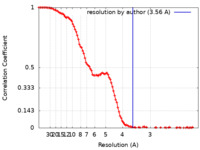
| Method | single particle reconstruction / cryo EM / Resolution: 3.56 Å | |||||||||
 Authors Authors | Blundell TL / Chaplin AK | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Using cryo-EM to understand antimycobacterial resistance in the catalase-peroxidase (KatG) from Mycobacterium tuberculosis. Authors: Asma Munir / Michael T Wilson / Steven W Hardwick / Dimitri Y Chirgadze / Jonathan A R Worrall / Tom L Blundell / Amanda K Chaplin /  Abstract: Resolution advances in cryoelectron microscopy (cryo-EM) now offer the possibility to visualize structural effects of naturally occurring resistance mutations in proteins and also of understanding ...Resolution advances in cryoelectron microscopy (cryo-EM) now offer the possibility to visualize structural effects of naturally occurring resistance mutations in proteins and also of understanding the binding mechanisms of small drug molecules. In Mycobacterium tuberculosis the multifunctional heme enzyme KatG is indispensable for activation of isoniazid (INH), a first-line pro-drug for treatment of tuberculosis. We present a cryo-EM methodology for structural and functional characterization of KatG and INH resistance variants. The cryo-EM structure of the 161 kDa KatG dimer in the presence of INH is reported to 2.7 Å resolution allowing the observation of potential INH binding sites. In addition, cryo-EM structures of two INH resistance variants, identified from clinical isolates, W107R and T275P, are reported. In combination with electronic absorbance spectroscopy our cryo-EM approach reveals how these resistance variants cause disorder in the heme environment preventing heme uptake and retention, providing insight into INH resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11689.map.gz emd_11689.map.gz | 87.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11689-v30.xml emd-11689-v30.xml emd-11689.xml emd-11689.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11689_fsc.xml emd_11689_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_11689.png emd_11689.png | 5.2 KB | ||
| Filedesc metadata |  emd-11689.cif.gz emd-11689.cif.gz | 6 KB | ||
| Others |  emd_11689_half_map_1.map.gz emd_11689_half_map_1.map.gz emd_11689_half_map_2.map.gz emd_11689_half_map_2.map.gz | 86.1 MB 86.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11689 http://ftp.pdbj.org/pub/emdb/structures/EMD-11689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11689 | HTTPS FTP |
-Related structure data
| Related structure data |  7aa3MC  6zjiC  7a2iC  7a7aC  7a7cC  7a8zC  7ag8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11689.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11689.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_11689_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11689_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimer of KatG from M.tuberculosis
| Entire | Name: Homodimer of KatG from M.tuberculosis |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer of KatG from M.tuberculosis
| Supramolecule | Name: Homodimer of KatG from M.tuberculosis / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Catalase-peroxidase
| Macromolecule | Name: Catalase-peroxidase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: catalase-peroxidase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.683617 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPEQHPPITE TTTGAASNGC PVVGHMKYPV EGGGNQDWWP NRLNLKVLHQ NPAVADPMGA AFDYAAEVAT IDVDALTRDI EEVMTTSQP WWPADYGHYG PLFIRMAWHA AGTYRIHDGR GGAGGGMQRF APLNSWPDNA SLDKARRLLW PVKKKYGKKL S WADLIVFA ...String: MPEQHPPITE TTTGAASNGC PVVGHMKYPV EGGGNQDWWP NRLNLKVLHQ NPAVADPMGA AFDYAAEVAT IDVDALTRDI EEVMTTSQP WWPADYGHYG PLFIRMAWHA AGTYRIHDGR GGAGGGMQRF APLNSWPDNA SLDKARRLLW PVKKKYGKKL S WADLIVFA GNCALESMGF KTFGFGFGRV DQWEPDEVYW GKEATWLGDE RYSGKRDLEN PLAAVQMGLI YVNPEGPNGN PD PMAAAVD IRETFRRMAM NDVETAALIV GGHTFGKPHG AGPADLVGPE PEAAPLEQMG LGWKSSYGTG TGKDAITSGI EVV WTNTPT KWDNSFLEIL YGYEWELTKS PAGAWQYTAK DGAGAGTIPD PFGGPGRSPT MLATDLSLRV DPIYERITRR WLEH PEELA DEFAKAWYKL IHRDMGPVAR YLGPLVPKQT LLWQDPVPAV SHDLVGEAEI ASLKSQIRAS GLTVSQLVST AWAAA SSFR GSDKRGGANG GRIRLQPQVG WEVNDPDGDL RKVIRTLEEI QESFNSAAPG NIKVSFADLV VLGGCAAIEK AAKAAG HNI TVPFTPGRTD ASQEQTDVES FAVLEPKADG FRNYLGKGNP LPAEYMLLDK ANLLTLSAPE MTVLVGGLRV LGANYKR LP LGVFTEASES LTNDFFVNLL DMGITWEPSP ADDGTYQGKD GSGKVKWTGS RVDLVFGSNS ELRALVEVYG ADDAQPKF V QDFVAAWDKV MNLDRFDVR UniProtKB: Catalase-peroxidase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)