[English] 日本語
 Yorodumi
Yorodumi- EMDB-11515: Molecular pore spanning the double membranes of a mutant murine c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11515 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular pore spanning the double membranes of a mutant murine coronaviral (MHV-A59 GFP-nsp3) replication organelles. | |||||||||
 Map data Map data | In situ structure of the molecular pore spanning the double-membranes of coronavirus mutant (MHV-A59 delta2-GFP3) induced replication organelles. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Murine hepatitis virus strain A59 Murine hepatitis virus strain A59 | |||||||||
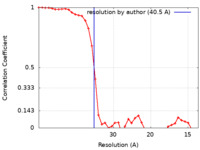
| Method | subtomogram averaging / cryo EM / Resolution: 40.5 Å | |||||||||
 Authors Authors | Barcena M / Wolff G | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: A molecular pore spans the double membrane of the coronavirus replication organelle. Authors: Georg Wolff / Ronald W A L Limpens / Jessika C Zevenhoven-Dobbe / Ulrike Laugks / Shawn Zheng / Anja W M de Jong / Roman I Koning / David A Agard / Kay Grünewald / Abraham J Koster / Eric J ...Authors: Georg Wolff / Ronald W A L Limpens / Jessika C Zevenhoven-Dobbe / Ulrike Laugks / Shawn Zheng / Anja W M de Jong / Roman I Koning / David A Agard / Kay Grünewald / Abraham J Koster / Eric J Snijder / Montserrat Bárcena /    Abstract: Coronavirus genome replication is associated with virus-induced cytosolic double-membrane vesicles, which may provide a tailored microenvironment for viral RNA synthesis in the infected cell. ...Coronavirus genome replication is associated with virus-induced cytosolic double-membrane vesicles, which may provide a tailored microenvironment for viral RNA synthesis in the infected cell. However, it is unclear how newly synthesized genomes and messenger RNAs can travel from these sealed replication compartments to the cytosol to ensure their translation and the assembly of progeny virions. In this study, we used cellular cryo-electron microscopy to visualize a molecular pore complex that spans both membranes of the double-membrane vesicle and would allow export of RNA to the cytosol. A hexameric assembly of a large viral transmembrane protein was found to form the core of the crown-shaped complex. This coronavirus-specific structure likely plays a key role in coronavirus replication and thus constitutes a potential drug target. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11515.map.gz emd_11515.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11515-v30.xml emd-11515-v30.xml emd-11515.xml emd-11515.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11515_fsc.xml emd_11515_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_11515.png emd_11515.png | 61.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11515 http://ftp.pdbj.org/pub/emdb/structures/EMD-11515 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11515 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11515 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11515.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11515.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In situ structure of the molecular pore spanning the double-membranes of coronavirus mutant (MHV-A59 delta2-GFP3) induced replication organelles. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Molecular pore containing hexameric nsp3 in the double membranes ...
| Entire | Name: Molecular pore containing hexameric nsp3 in the double membranes of the replication organelles induced by murine coronavirus mutant (MHV-A59, delta2-GFP3 mutant). |
|---|---|
| Components |
|
-Supramolecule #1: Molecular pore containing hexameric nsp3 in the double membranes ...
| Supramolecule | Name: Molecular pore containing hexameric nsp3 in the double membranes of the replication organelles induced by murine coronavirus mutant (MHV-A59, delta2-GFP3 mutant). type: complex / ID: 1 / Parent: 0 Details: Upon infection with murine coronavirus (MHV-A59 delta2-GFP3, doi: 10.1128/JVI.00021-14) the formation of ER derived double-membrane vesicles is induced in the host cell. The molecular pore ...Details: Upon infection with murine coronavirus (MHV-A59 delta2-GFP3, doi: 10.1128/JVI.00021-14) the formation of ER derived double-membrane vesicles is induced in the host cell. The molecular pore spans the two membranes of these viral replication organelles. |
|---|---|
| Source (natural) | Organism:  Murine hepatitis virus strain A59 / Organelle: virus-induced double-membrane vesicles / Location in cell: perinuclear area Murine hepatitis virus strain A59 / Organelle: virus-induced double-membrane vesicles / Location in cell: perinuclear area |
| Recombinant expression | Organism:  Murine hepatitis virus strain A59 Murine hepatitis virus strain A59 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 310 K / Instrument: LEICA EM GP / Details: Blotting time 15s. |
| Details | electron cryo-tomography of FIB-milled lamellae |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)