[English] 日本語
 Yorodumi
Yorodumi- EMDB-11496: Subtomogram averaging of SARS-CoV-2 spike protein from unconcentr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11496 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram averaging of SARS-CoV-2 spike protein from unconcentrated virions: Prefusion Class (2 open RBDs) | ||||||||||||||||||
 Map data Map data | prefusion 2 open RBDs | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 9.9 Å | ||||||||||||||||||
 Authors Authors | Ke Z / Oton J / Zivanov J / Lu JM / Peukes J / Cortese M / Zila V / Scheres SHW / Briggs JAG | ||||||||||||||||||
| Funding support | European Union,  United Kingdom, United Kingdom,  Japan, Japan,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Authors: Zunlong Ke / Joaquin Oton / Kun Qu / Mirko Cortese / Vojtech Zila / Lesley McKeane / Takanori Nakane / Jasenko Zivanov / Christopher J Neufeldt / Berati Cerikan / John M Lu / Julia Peukes / ...Authors: Zunlong Ke / Joaquin Oton / Kun Qu / Mirko Cortese / Vojtech Zila / Lesley McKeane / Takanori Nakane / Jasenko Zivanov / Christopher J Neufeldt / Berati Cerikan / John M Lu / Julia Peukes / Xiaoli Xiong / Hans-Georg Kräusslich / Sjors H W Scheres / Ralf Bartenschlager / John A G Briggs /   Abstract: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virions are surrounded by a lipid bilayer from which spike (S) protein trimers protrude. Heavily glycosylated S trimers bind to the ...Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virions are surrounded by a lipid bilayer from which spike (S) protein trimers protrude. Heavily glycosylated S trimers bind to the angiotensin-converting enzyme 2 receptor and mediate entry of virions into target cells. S exhibits extensive conformational flexibility: it modulates exposure of its receptor-binding site and subsequently undergoes complete structural rearrangement to drive fusion of viral and cellular membranes. The structures and conformations of soluble, overexpressed, purified S proteins have been studied in detail using cryo-electron microscopy, but the structure and distribution of S on the virion surface remain unknown. Here we applied cryo-electron microscopy and tomography to image intact SARS-CoV-2 virions and determine the high-resolution structure, conformational flexibility and distribution of S trimers in situ on the virion surface. These results reveal the conformations of S on the virion, and provide a basis from which to understand interactions between S and neutralizing antibodies during infection or vaccination. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11496.map.gz emd_11496.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11496-v30.xml emd-11496-v30.xml emd-11496.xml emd-11496.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11496.png emd_11496.png | 35.5 KB | ||
| Others |  emd_11496_half_map_1.map.gz emd_11496_half_map_1.map.gz emd_11496_half_map_2.map.gz emd_11496_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11496 http://ftp.pdbj.org/pub/emdb/structures/EMD-11496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11496 | HTTPS FTP |
-Related structure data
| Related structure data |  6zwvC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10493 (Title: Subtomogram averaging and classification of SARS-CoV-2 Spike Proteins on intact virions EMPIAR-10493 (Title: Subtomogram averaging and classification of SARS-CoV-2 Spike Proteins on intact virionsData size: 372.4 Data #1: Unaligned Frames from the microscope. Data were recorded [tilt series] Data #2: Aligned and order sorted stack of the tilt series. This is not dose-filtered stack. Associated mdoc files are included in this folder. [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11496.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11496.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | prefusion 2 open RBDs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.064 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: prefusion 2 open RBDs, half1
| File | emd_11496_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | prefusion 2 open RBDs, half1 | ||||||||||||
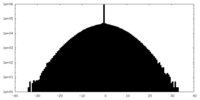
| Projections & Slices |
| ||||||||||||
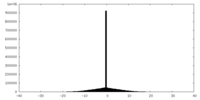
| Density Histograms |
-Half map: prefusion 2 open RBDs, half2
| File | emd_11496_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | prefusion 2 open RBDs, half2 | ||||||||||||
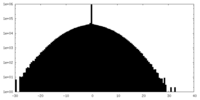
| Projections & Slices |
| ||||||||||||
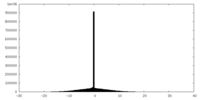
| Density Histograms |
- Sample components
Sample components
-Entire : Severe acute respiratory syndrome coronavirus 2
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Severe acute respiratory syndrome coronavirus 2
| Supramolecule | Name: Severe acute respiratory syndrome coronavirus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Unconcentrated supernatant of SARS-CoV-2 virions produced from infected VeroE6 cells. NCBI-ID: 2697049 Sci species name: Severe acute respiratory syndrome coronavirus 2 Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: SARS-CoV-2 Spike proteins on virions
| Macromolecule | Name: SARS-CoV-2 Spike proteins on virions / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQGVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PRRARSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GAALQIPFAM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TASALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDKVEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE E(NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG) (NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG)(NAG) (NAG)(NAG)LDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQELGKYEQY IKWPWYIWLG FIAGLIAIVM VTIMLCCMTS CCSCLKGCCS CGSCCKFDED DSEPVLKGVK LHYT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-2/2 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-10 / Average exposure time: 1.25 sec. / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)