[English] 日本語
 Yorodumi
Yorodumi- EMDB-1142: Localization of the coactivator Cdh1 and the cullin subunit Apc2 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1142 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. | |||||||||
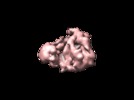
 Map data Map data | This is a 3D map of the Xenopus APC/C in complex with an activator Cdh1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Dube P / Herzog F / Peters JM / Stark H | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2005 Journal: Mol Cell / Year: 2005Title: Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Authors: Prakash Dube / Franz Herzog / Christian Gieffers / Bjoern Sander / Dietmar Riedel / Shirley A Müller / Andreas Engel / Jan-Michael Peters / Holger Stark /  Abstract: The anaphase-promoting complex/cyclosome (APC/C) is a ubiquitin ligase with essential functions in mitosis, meiosis, and G1 phase of the cell cycle. APC/C recognizes substrates via coactivator ...The anaphase-promoting complex/cyclosome (APC/C) is a ubiquitin ligase with essential functions in mitosis, meiosis, and G1 phase of the cell cycle. APC/C recognizes substrates via coactivator proteins such as Cdh1, and bound substrates are ubiquitinated by E2 enzymes that interact with a hetero-dimer of the RING subunit Apc11 and the cullin Apc2. We have obtained three-dimensional (3D) models of human and Xenopus APC/C by angular reconstitution and random conical tilt (RCT) analyses of negatively stained cryo-electron microscopy (cryo-EM) preparations, have determined the masses of these particles by scanning transmission electron microscopy (STEM), and have mapped the locations of Cdh1 and Apc2. These proteins are located on the same side of the asymmetric APC/C, implying that this is where substrates are ubiquitinated. We have further identified a large flexible domain in APC/C that adopts a different orientation upon Cdh1 binding. Cdh1 may thus activate APC/C both by recruiting substrates and by inducing conformational changes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1142.map.gz emd_1142.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1142-v30.xml emd-1142-v30.xml emd-1142.xml emd-1142.xml | 8.1 KB 8.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1142.gif 1142.gif | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1142 http://ftp.pdbj.org/pub/emdb/structures/EMD-1142 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1142 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1142 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1142.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1142.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a 3D map of the Xenopus APC/C in complex with an activator Cdh1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.318 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Anaphase Promoting Complex
| Entire | Name: Anaphase Promoting Complex |
|---|---|
| Components |
|
-Supramolecule #1000: Anaphase Promoting Complex
| Supramolecule | Name: Anaphase Promoting Complex / type: sample / ID: 1000 / Oligomeric state: Monomer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 1.5 MDa |
-Macromolecule #1: Anaphase Promoting Complex
| Macromolecule | Name: Anaphase Promoting Complex / type: protein_or_peptide / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #2: Cdh1
| Macromolecule | Name: Cdh1 / type: protein_or_peptide / ID: 2 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 / Details: 20 mM Hepes, 150 mM NaCl, 0.5 mM DTT |
|---|---|
| Staining | Type: NEGATIVE / Details: carbon sandwich with 2% x/v uranyl formate |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Average electron dose: 15 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 90500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal magnification: 90500 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name: Imagic-5 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















