[English] 日本語
 Yorodumi
Yorodumi- EMDB-11106: Poliovirus type 3 (strain Saukett) stabilised virus-like particle... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11106 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) from a mammalian expression system. | |||||||||
 Map data Map data | MRC map filtered after local resolution in RELION. Recommended contour level 0.07 after inspection in Chimera. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid protein / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcaveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane ...caveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / host cell cytoplasm / DNA replication / RNA helicase activity / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Human poliovirus 3 Human poliovirus 3 | |||||||||
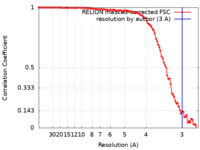
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Bahar MW / Porta C | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: NPJ Vaccines / Year: 2021 Journal: NPJ Vaccines / Year: 2021Title: Mammalian expression of virus-like particles as a proof of principle for next generation polio vaccines. Authors: Mohammad W Bahar / Claudine Porta / Helen Fox / Andrew J Macadam / Elizabeth E Fry / David I Stuart /  Abstract: Global vaccination programs using live-attenuated oral and inactivated polio vaccine (OPV and IPV) have almost eradicated poliovirus (PV) but these vaccines or their production pose significant risk ...Global vaccination programs using live-attenuated oral and inactivated polio vaccine (OPV and IPV) have almost eradicated poliovirus (PV) but these vaccines or their production pose significant risk in a polio-free world. Recombinant PV virus-like particles (VLPs), lacking the viral genome, represent safe next-generation vaccines, however their production requires optimisation. Here we present an efficient mammalian expression strategy producing good yields of wild-type PV VLPs for all three serotypes and a thermostabilised variant for PV3. Whilst the wild-type VLPs were predominantly in the non-native C-antigenic form, the thermostabilised PV3 VLPs adopted the native D-antigenic conformation eliciting neutralising antibody titres equivalent to the current IPV and were indistinguishable from natural empty particles by cryo-electron microscopy with a similar stabilising lipidic pocket-factor in the VP1 β-barrel. This factor may not be available in alternative expression systems, which may require synthetic pocket-binding factors. VLPs equivalent to these mammalian expressed thermostabilized particles, represent safer non-infectious vaccine candidates for the post-eradication era. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11106.map.gz emd_11106.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11106-v30.xml emd-11106-v30.xml emd-11106.xml emd-11106.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11106_fsc.xml emd_11106_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11106.png emd_11106.png | 331.3 KB | ||
| Filedesc metadata |  emd-11106.cif.gz emd-11106.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11106 http://ftp.pdbj.org/pub/emdb/structures/EMD-11106 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11106 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11106 | HTTPS FTP |
-Related structure data
| Related structure data |  6z6wMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11106.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11106.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
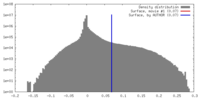
| Annotation | MRC map filtered after local resolution in RELION. Recommended contour level 0.07 after inspection in Chimera. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human poliovirus 3
| Entire | Name:  Human poliovirus 3 Human poliovirus 3 |
|---|---|
| Components |
|
-Supramolecule #1: Human poliovirus 3
| Supramolecule | Name: Human poliovirus 3 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Recombinantly expressed virus-like particle of PV3 (strain Saukett). NCBI-ID: 12086 / Sci species name: Human poliovirus 3 / Sci species strain: Saukett / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.85 MDa |
| Virus shell | Shell ID: 1 / Name: Virus shell 1 / Diameter: 310.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid proteins, VP1
| Macromolecule | Name: Capsid proteins, VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 Human poliovirus 3 |
| Molecular weight | Theoretical: 33.562785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIEDLITEVA QGALTLSLPK QQDSLPDTKA SGPAHSKEVP ALTAVETGAT NPLVPSDTVQ TRHVIQRRSR SESTIESFFA RGACVAIIE VDNEEPTTRA QKLFAMWRIT YKDTVQLRRK LEFFTYSRFD MELTFVVTAN FTNTNNGHAL NQVYQIMYIP P GAPTPKSW ...String: GIEDLITEVA QGALTLSLPK QQDSLPDTKA SGPAHSKEVP ALTAVETGAT NPLVPSDTVQ TRHVIQRRSR SESTIESFFA RGACVAIIE VDNEEPTTRA QKLFAMWRIT YKDTVQLRRK LEFFTYSRFD MELTFVVTAN FTNTNNGHAL NQVYQIMYIP P GAPTPKSW DDYTWQTSSN PSIFYTYGAA PARISVPYVG LANAYSHFYD GFAKVPLKTD ANDQIGDSLY SAMTVDDFGV LA IRVVNDH NPTKVTSKVR IYMKPKHVRV WCPRPPRAVP YYGPGVDYKD NLNPLSEKGL TTY UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid proteins, VP0
| Macromolecule | Name: Capsid proteins, VP0 / type: protein_or_peptide / ID: 2 Details: Sequence is given for the VP0 polypeptide. Mutations are numbered according to sequence numbering for mature polypeptides VP2 and VP4. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 Human poliovirus 3 |
| Molecular weight | Theoretical: 37.623023 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGAQVSSQKV GAHENSNRAY GGSTINYTTI NYYKDSASNA ASKQDYSQDP SKFTEPLKDV LIKTAPALNS PNVEACGYSD RVLQLTIGN STITTQEAAN SVVAYGRWPE FIRDDEANPV DQPTEPDVAT CRFYTLDTVM WGKESKGWWW KLPDALRDMG L FGQNMYYH ...String: MGAQVSSQKV GAHENSNRAY GGSTINYTTI NYYKDSASNA ASKQDYSQDP SKFTEPLKDV LIKTAPALNS PNVEACGYSD RVLQLTIGN STITTQEAAN SVVAYGRWPE FIRDDEANPV DQPTEPDVAT CRFYTLDTVM WGKESKGWWW KLPDALRDMG L FGQNMYYH YLGRSGYTVH VQCNASKFHQ GALGVFAIPE YCLAGDSDKQ RYTSYANANP GEKGGKFYSQ FNRDTAVTSP KR EFCPVDY LLGCGVLLGN AFVYPHQIIN LRTNNSATIV LPYVNAMAID SMVKHNNWGI AILPLSPLDF AQESSVEIPI TVT IAPMCS EFNGLRNVTA PKFQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid proteins, VP3
| Macromolecule | Name: Capsid proteins, VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 Human poliovirus 3 |
| Molecular weight | Theoretical: 26.3151 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GLPVLNTPGS NQYLTSDNYQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLENT KRNTMDMYRV TLSDSADLSQ PILCFSLSP ASDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV ...String: GLPVLNTPGS NQYLTSDNYQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLENT KRNTMDMYRV TLSDSADLSQ PILCFSLSP ASDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV PWISNVTYRQ TTQDSFTEGG YISMFYQTRI VVPLSTPKSM SMLGFVSACN DFSVRLLRDT THISQSALPQ UniProtKB: Genome polyprotein |
-Macromolecule #4: SPHINGOSINE
| Macromolecule | Name: SPHINGOSINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: SPH |
|---|---|
| Molecular weight | Theoretical: 299.492 Da |
| Chemical component information |  ChemComp-SPH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 / Component:
| ||||||
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR Details: The specific type of grid used was Ultra-thin carbon support film, 3nm - on lacey carbon AGS187-4 from Agar Scientific. | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Double blotting with 4 ul of sample, followed by 4 second blot, before plunging.. | ||||||
| Details | Recombinantly expressed VLP of PV3. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-25 / Number grids imaged: 2 / Number real images: 1465 / Average exposure time: 5.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 160000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial model was rigid body fitted using UCSF chimera, and the refined in real space using Phenix_real.space.refine. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-6z6w: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)