[English] 日本語
 Yorodumi
Yorodumi- EMDB-11082: Cryo-EM structure of the A. baumannii MlaBDEF complex bound to APPNHP -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11082 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the A. baumannii MlaBDEF complex bound to APPNHP | |||||||||
 Map data Map data | Map of the A. baumannii MlaBDEF complex, bound to APPNHP, at 3.9A resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | lipid transport / antibiotic resistance / ABC transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphospholipid transporter activity / ATP-binding cassette (ABC) transporter complex / phospholipid binding / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Mann D / Bergeron JRC | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Structure and lipid dynamics in the maintenance of lipid asymmetry inner membrane complex of A. baumannii. Authors: Daniel Mann / Junping Fan / Kamolrat Somboon / Daniel P Farrell / Andrew Muenks / Svetomir B Tzokov / Frank DiMaio / Syma Khalid / Samuel I Miller / Julien R C Bergeron /     Abstract: Multi-resistant bacteria are a major threat in modern medicine. The gram-negative coccobacillus Acinetobacter baumannii currently leads the WHO list of pathogens in critical need for new therapeutic ...Multi-resistant bacteria are a major threat in modern medicine. The gram-negative coccobacillus Acinetobacter baumannii currently leads the WHO list of pathogens in critical need for new therapeutic development. The maintenance of lipid asymmetry (MLA) protein complex is one of the core machineries that transport lipids from/to the outer membrane in gram-negative bacteria. It also contributes to broad-range antibiotic resistance in several pathogens, most prominently in A. baumannii. Nonetheless, the molecular details of its role in lipid transport has remained largely elusive. Here, we report the cryo-EM maps of the core MLA complex, MlaBDEF, from the pathogen A. baumannii, in the apo-, ATP- and ADP-bound states, revealing multiple lipid binding sites in the cytosolic and periplasmic side of the complex. Molecular dynamics simulations suggest their potential trajectory across the membrane. Collectively with the recently-reported structures of the E. coli orthologue, this data also allows us to propose a molecular mechanism of lipid transport by the MLA system. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Structure and lipid dynamics in the A. baumannii maintenance of lipid asymmetry (MLA) inner membrane complex Authors: Mann D / Fan J / Farrell DP / Somboon K / Muenks A / Tzokov SB / Khalid S / Dimaio F / Miller SI / Bergeron JRC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11082.map.gz emd_11082.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11082-v30.xml emd-11082-v30.xml emd-11082.xml emd-11082.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11082.png emd_11082.png | 168.6 KB | ||
| Masks |  emd_11082_msk_1.map emd_11082_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11082.cif.gz emd-11082.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11082 http://ftp.pdbj.org/pub/emdb/structures/EMD-11082 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11082 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11082 | HTTPS FTP |
-Related structure data
| Related structure data |  6z5uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10425 (Title: Cryo-EM structure of the A.baumannii MlaBDEF complex bound to AppNHp EMPIAR-10425 (Title: Cryo-EM structure of the A.baumannii MlaBDEF complex bound to AppNHpData size: 135.6 Data #1: MotionCor2 aligned micrographs [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11082.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11082.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the A. baumannii MlaBDEF complex, bound to APPNHP, at 3.9A resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
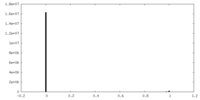
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11082_msk_1.map emd_11082_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
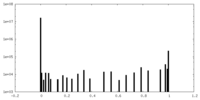
| Density Histograms |
- Sample components
Sample components
-Entire : MlaBDEF complex
| Entire | Name: MlaBDEF complex |
|---|---|
| Components |
|
-Supramolecule #1: MlaBDEF complex
| Supramolecule | Name: MlaBDEF complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
-Macromolecule #1: ABC transporter permease
| Macromolecule | Name: ABC transporter permease / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 27.322443 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNTIAWLGRL VIERIRGIGV AALMLLQIIF SLPSAGGFGR FVYQMHRVGV MSLLIITVSG LFIGLVLGLQ GYSILVNVGS ESMLGTMVS LTLLRELAPV VAALLFAGRA GSALTAEIGS MKQSEQLASM EMIGVDPLKQ IVSPRLWAGI VSLPMLTVIF A AIGIVGGK ...String: MNTIAWLGRL VIERIRGIGV AALMLLQIIF SLPSAGGFGR FVYQMHRVGV MSLLIITVSG LFIGLVLGLQ GYSILVNVGS ESMLGTMVS LTLLRELAPV VAALLFAGRA GSALTAEIGS MKQSEQLASM EMIGVDPLKQ IVSPRLWAGI VSLPMLTVIF A AIGIVGGK LVGVDFLGVD EGSFWSGMQN NVQFGHDVVN GIIKSIVFAL LCTWIAVFQG YACDPTPEGI ATAMTRTVVY SS LCVLGFD FVLTAVMFGG I UniProtKB: Intermembrane phospholipid transport system permease protein MlaE |
-Macromolecule #2: Anti-sigma factor antagonist
| Macromolecule | Name: Anti-sigma factor antagonist / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 10.874672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVQYLNQELV VSGKIDFENA EQQYQAGLAI IKKQTSFPLI VDLKQLEHGN TLALAVLVQW LRQTPQKSGL HFKNVPEKML KIIQACHLQ EDLHLV UniProtKB: Anti-sigma factor antagonist |
-Macromolecule #3: MCE family protein
| Macromolecule | Name: MCE family protein / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 24.135322 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSRTSELAV GIFVIIFGIA LFFLAMKVSG LVGTNLSDGY TMKAQFDNVN GLKPRAKVTM SGVTIGRVDS ITLDPVTRLA TVTFDLDGK LTSFNAEQLK EVQKNALDEL RYSSDYTQAT PAQQKTMEQQ LISNMNSITS IDEDAYIMVA TNGLLGEKYL K IVPGGGLN ...String: MKSRTSELAV GIFVIIFGIA LFFLAMKVSG LVGTNLSDGY TMKAQFDNVN GLKPRAKVTM SGVTIGRVDS ITLDPVTRLA TVTFDLDGK LTSFNAEQLK EVQKNALDEL RYSSDYTQAT PAQQKTMEQQ LISNMNSITS IDEDAYIMVA TNGLLGEKYL K IVPGGGLN YLKRGDTISN TQGTMDLEDL ISKFITGGGA GKVAAGSSSA EEKAPASTDS SAQPSFVE UniProtKB: ABC transporter periplasmic substrate-binding protein |
-Macromolecule #4: ABC transporter ATP-binding protein
| Macromolecule | Name: ABC transporter ATP-binding protein / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 30.367848 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIAIMNNKTP LSTQSLIEVK NLSFNRGERV IYDNISLNIR RGQITAIMGP SGTGKTTLLR LIGGQLVPDQ GEVLLDGKDI AQMSRQELF AARARMGMLF QSGALFTDMS VYENVAFPIR AHTKLSENLI AELVALKLES VGLRGTEQLM PTELSGGMNR R VALARAIA ...String: MIAIMNNKTP LSTQSLIEVK NLSFNRGERV IYDNISLNIR RGQITAIMGP SGTGKTTLLR LIGGQLVPDQ GEVLLDGKDI AQMSRQELF AARARMGMLF QSGALFTDMS VYENVAFPIR AHTKLSENLI AELVALKLES VGLRGTEQLM PTELSGGMNR R VALARAIA LDPDLIMYDE PFAGQDPIVK GVLTRLIRSL REALDLTTII VSHDVPETLS IADYIYVVAE GKIQGEGTPE EL QAYASPF VKQFLTGSAE GPVEYQFSHQ AYLDNEVRP UniProtKB: ABC transporter ATP-binding protein |
-Macromolecule #5: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 93295 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)