+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11008 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Icosahedral reconstruction of human adenovirus D26 capsid | |||||||||
 Map data Map data | Icosahedral map of human adenovirus D26 capsid | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
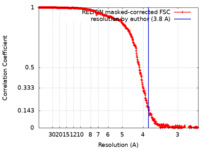
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Huiskonen JT / Abrishami V | |||||||||
| Funding support | European Union,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Prog Biophys Mol Biol / Year: 2021 Journal: Prog Biophys Mol Biol / Year: 2021Title: Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Authors: Vahid Abrishami / Serban L Ilca / Josue Gomez-Blanco / Ilona Rissanen / José Miguel de la Rosa-Trevín / Vijay S Reddy / José-Maria Carazo / Juha T Huiskonen /       Abstract: Technological advances in transmission electron microscopes and detectors have turned cryogenic electron microscopy (cryo-EM) into an essential tool for structural biology. A commonly used cryo-EM ...Technological advances in transmission electron microscopes and detectors have turned cryogenic electron microscopy (cryo-EM) into an essential tool for structural biology. A commonly used cryo-EM data analysis method, single particle analysis, averages hundreds of thousands of low-dose images of individual macromolecular complexes to determine a density map of the complex. The presence of symmetry in the complex is beneficial since each projection image can be assigned to multiple views of the complex. However, data processing that applies symmetry can average out asymmetric features and consequently data analysis methods are required to resolve asymmetric structural features. Scipion is a cryo-EM image processing framework that integrates functions from different image processing packages as plugins. To extend its functionality for handling symmetry mismatches, we present here a Scipion plugin termed LocalRec implementing the localized reconstruction method. When tested on an adenovirus data set, the plugin enables resolving the symmetry-mismatched trimeric fibre bound to the five-fold vertices of the capsid. Furthermore, it improves the structure determination of the icosahedral capsid by dealing with the defocus gradient across the particle. LocalRec is expected to be widely applicable in a range of cryo-EM investigations of flexible and symmetry mismatched complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11008.map.gz emd_11008.map.gz | 5.9 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11008-v30.xml emd-11008-v30.xml emd-11008.xml emd-11008.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11008_fsc.xml emd_11008_fsc.xml | 42.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11008.png emd_11008.png | 211.4 KB | ||
| Masks |  emd_11008_msk_1.map emd_11008_msk_1.map | 6.4 GB |  Mask map Mask map | |
| Others |  emd_11008_additional.map.gz emd_11008_additional.map.gz emd_11008_additional_1.map.gz emd_11008_additional_1.map.gz emd_11008_half_map_1.map.gz emd_11008_half_map_1.map.gz emd_11008_half_map_2.map.gz emd_11008_half_map_2.map.gz | 999.2 MB 999.2 MB 5.3 GB 5.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11008 http://ftp.pdbj.org/pub/emdb/structures/EMD-11008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11008 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10455 (Title: Single particle cryo-EM dataset of human adenovirus HAdV-D26 EMPIAR-10455 (Title: Single particle cryo-EM dataset of human adenovirus HAdV-D26Data size: 46.5 Data #1: Particle stacks of human adenovirus HAdV-D26 [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11008.map.gz / Format: CCP4 / Size: 6.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11008.map.gz / Format: CCP4 / Size: 6.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral map of human adenovirus D26 capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11008_msk_1.map emd_11008_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
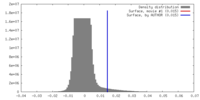
| Density Histograms |
-Additional map: #1
| File | emd_11008_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
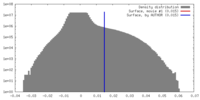
| Density Histograms |
-Additional map: #1
| File | emd_11008_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half 1 of human adenovirus D26 capsid map
| File | emd_11008_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 1 of human adenovirus D26 capsid map | ||||||||||||
| Projections & Slices |
| ||||||||||||
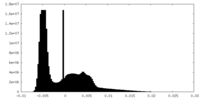
| Density Histograms |
-Half map: Half 2 of human adenovirus D26 capsid map
| File | emd_11008_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 2 of human adenovirus D26 capsid map | ||||||||||||
| Projections & Slices |
| ||||||||||||
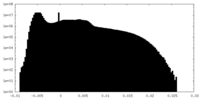
| Density Histograms |
- Sample components
Sample components
-Entire : Icosahedral reconstruction of human adenovirus D26
| Entire | Name: Icosahedral reconstruction of human adenovirus D26 |
|---|---|
| Components |
|
-Supramolecule #1: Icosahedral reconstruction of human adenovirus D26
| Supramolecule | Name: Icosahedral reconstruction of human adenovirus D26 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
-Supramolecule #2: A composite volume of human adenovirus D26 capsid created from lo...
| Supramolecule | Name: A composite volume of human adenovirus D26 capsid created from localized reconstructions of the four hexons type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.1 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 7-38 / Number real images: 2000 / Average exposure time: 7.6 sec. / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)