[English] 日本語
 Yorodumi
Yorodumi- EMDB-1096: Major conformational change in the complex SF3b upon integration ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1096 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. | |||||||||
 Map data Map data | Volume file of the native human U11/U12 di-snRNP | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.4 Å | |||||||||
 Authors Authors | Golas MM / Sander B / Will CL / Luhrmann R | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2005 Journal: Mol Cell / Year: 2005Title: Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Authors: Monika M Golas / Bjoern Sander / Cindy L Will / Reinhard Lührmann / Holger Stark /  Abstract: In some eukaryotes, a minor class of introns is removed by the U12-dependent spliceosome, which contains the small nuclear ribonucleoprotein (snRNP) heterodimer U11/U12. The U11/U12 di-snRNP forms a ...In some eukaryotes, a minor class of introns is removed by the U12-dependent spliceosome, which contains the small nuclear ribonucleoprotein (snRNP) heterodimer U11/U12. The U11/U12 di-snRNP forms a molecular bridge that functionally pairs the intron ends of the pre-mRNA. We have determined the three-dimensional (3D) structure of the human U11/U12 di-snRNP by single particle electron cryomicroscopy using angular reconstitution and random conical tilt. SF3b, a heteromeric protein complex functionally important for branch site recognition, was located in the U11/U12 di-snRNP by antibody labeling and by identification of structural domains of SF3b155, SF3b49, and p14. The conformation of SF3b bound to the U11/U12 di-snRNP differs from that of isolated SF3b: upon integration into the di-snRNP, SF3b rearranges into a more open form. The manner in which SF3b is integrated in the U11/U12 di-snRNP has important implications for branch site recognition. Furthermore, a putative model of the pre-mRNA binding to the U11/U12 di-snRNP is proposed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1096.map.gz emd_1096.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1096-v30.xml emd-1096-v30.xml emd-1096.xml emd-1096.xml | 6.1 KB 6.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1096.gif 1096.gif | 15.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1096 http://ftp.pdbj.org/pub/emdb/structures/EMD-1096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1096 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1096.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1096.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Volume file of the native human U11/U12 di-snRNP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human native U11U12 di-snRNP
| Entire | Name: human native U11U12 di-snRNP |
|---|---|
| Components |
|
-Supramolecule #1000: human native U11U12 di-snRNP
| Supramolecule | Name: human native U11U12 di-snRNP / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 1.3 MDa |
-Supramolecule #1: U11U12 di-snRNP
| Supramolecule | Name: U11U12 di-snRNP / type: organelle_or_cellular_component / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: NITROGEN |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Electron beam | Acceleration voltage: 160 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Gatan / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 13.4 Å / Resolution method: FSC 0.5 CUT-OFF |
|---|
 Movie
Movie Controller
Controller


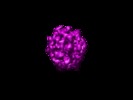








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















