[English] 日本語
 Yorodumi
Yorodumi- EMDB-10827: Negative stain reconstruction of grafix crosslink human condensin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10827 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain reconstruction of grafix crosslink human condensin I in the presence of ATPyS | |||||||||
 Map data Map data | Human condensin I negative stain reconstruction, crosslinked with glutaraldehyde in the presence of ATPyS | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
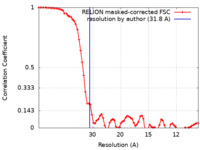
| Method | single particle reconstruction / negative staining / Resolution: 31.8 Å | |||||||||
 Authors Authors | Cutts EE / Beuron F / Morris E / Vannini A | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Human Condensin I and II Drive Extensive ATP-Dependent Compaction of Nucleosome-Bound DNA. Authors: Muwen Kong / Erin E Cutts / Dongqing Pan / Fabienne Beuron / Thangavelu Kaliyappan / Chaoyou Xue / Edward P Morris / Andrea Musacchio / Alessandro Vannini / Eric C Greene /     Abstract: Structural maintenance of chromosomes (SMC) complexes are essential for genome organization from bacteria to humans, but their mechanisms of action remain poorly understood. Here, we characterize ...Structural maintenance of chromosomes (SMC) complexes are essential for genome organization from bacteria to humans, but their mechanisms of action remain poorly understood. Here, we characterize human SMC complexes condensin I and II and unveil the architecture of the human condensin II complex, revealing two putative DNA-entrapment sites. Using single-molecule imaging, we demonstrate that both condensin I and II exhibit ATP-dependent motor activity and promote extensive and reversible compaction of double-stranded DNA. Nucleosomes are incorporated into DNA loops during compaction without being displaced from the DNA, indicating that condensin complexes can readily act upon nucleosome-bound DNA molecules. These observations shed light on critical processes involved in genome organization in human cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10827.map.gz emd_10827.map.gz | 988.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10827-v30.xml emd-10827-v30.xml emd-10827.xml emd-10827.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10827_fsc.xml emd_10827_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10827.png emd_10827.png | 35.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10827 http://ftp.pdbj.org/pub/emdb/structures/EMD-10827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10827 | HTTPS FTP |
-Validation report
| Summary document |  emd_10827_validation.pdf.gz emd_10827_validation.pdf.gz | 203.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10827_full_validation.pdf.gz emd_10827_full_validation.pdf.gz | 202.8 KB | Display | |
| Data in XML |  emd_10827_validation.xml.gz emd_10827_validation.xml.gz | 9.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10827 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10827 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10827 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10827 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10827.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10827.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human condensin I negative stain reconstruction, crosslinked with glutaraldehyde in the presence of ATPyS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Condensin I pentameric complex of SMC2, SMC4, CapH, CapD3 and CapG
| Entire | Name: Condensin I pentameric complex of SMC2, SMC4, CapH, CapD3 and CapG |
|---|---|
| Components |
|
-Supramolecule #1: Condensin I pentameric complex of SMC2, SMC4, CapH, CapD3 and CapG
| Supramolecule | Name: Condensin I pentameric complex of SMC2, SMC4, CapH, CapD3 and CapG type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 643 KDa |
-Macromolecule #1: SMC2
| Macromolecule | Name: SMC2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: HIKSIILEGF KSYAQRTEV NGFDPLFNAI TGLNGSGKSN ILDSICFLL GISNLSQVRA S NLQDLVYK NGQAGITKAS VS ITFDNSD KKQSPLGFEV HDE ITVTRQ VVIGGRNKYL INGV NANNT RVQDLFCSVG LNVNN PHFL IMQGRITKVL NMKPPE ILS ...String: HIKSIILEGF KSYAQRTEV NGFDPLFNAI TGLNGSGKSN ILDSICFLL GISNLSQVRA S NLQDLVYK NGQAGITKAS VS ITFDNSD KKQSPLGFEV HDE ITVTRQ VVIGGRNKYL INGV NANNT RVQDLFCSVG LNVNN PHFL IMQGRITKVL NMKPPE ILS MIEEAAGTRM YEYKKIA AQ KTIEKKEAKL KEIKTILE E EITPTIQKLK EERSSYLEY QKVMREIEHL SRLYIAYQFL LAEDTKVRS AEELKEMQDK V IKLQEELS ENDKKIKALN HE IEELEKR KDKETGGILR SLE DALAEA QRVNTKSQSA FDLK KKNLA CEESKRKELE KNMVE DSKT LAAKEKEVKK ITDGLH ALQ EASNKDAEAL AAAQQHF NA VSAGLSSNED GAEATLAG Q MMACKNDISK AQTEAKQAQ MKLKHAQQEL KNKQAEVKKM DSGYRKDQE ALEAVKRLKE K LEAEMKKL NYEENKEESL LE KRRQLSR DIGRLKETYE ALL ARFPNL RFAYKDPEKN WNRN CVKGL VASLISVKDT SATTA LELV AGERLYNVVV DTEVTG KKL LERGELKRRY TIIPLNK IS ARCIAPETLR VAQNLVGP D NVHVALSLVE YKPELQKAM EFVFGTTFVC DNMDNAKKVA FDKRIMTRT VTLGGDVFDP H GTLSGGAR SQAASILTKF QE LKDVQDE LRIKENELRA LEE ELAGLK NTAEKYRQLK QQWE MKTEE ADLLQTKLQQ SSYHK QQEE LDALKKTIEE SEETLK NTK EIQRKAEEKY EVLENKM KN AEAERERELK DAQKKLDC A KTKADASSKK MKEKQQEVE AITLELEELK REHTSYKQQL EAVNEAIKS YESQIEVMAA E VAKNKESV NKAQEEVTKQ KE VITAQDT VIKAKYAEVA KHK EQNNDS QLKIKELDHN ISKH KREAE DGAAKVSKML KDYDW INAE RHLFGQPNSA YDFKTN NPK EAGQRLQKLQ EMKEKLG RN VNMRAMNVLT EAEERYND L MKKKRIVEND KSKILTTIE DLDQKKNQAL NIAWQKVNKD FGSIFSTLL PGANAMLAPP E GQTVLDGL EFKVALGNTW KE NLTELSG GQRSLVALSL ILS MLLFKP APIYILDEVD AALD LSHTQ NIGQMLRTHF THSQF IVVS LKEGMFNNAN VLFKTK FVD GVSTVARFTQ CQNGKIS KE AKSKAKPPKG AHVEV |
-Macromolecule #2: SMC4
| Macromolecule | Name: SMC4 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAHHHHHHHH HHLEVLFQGP RKGTQPSTA RRREEGPPPP S PDGASSDA EPEPPSGRTE SP ATAAETA SEELDNRSLE EIL NSIPPP PPPAMTNEAG APRL MITHI VNQNFKSYAG EKILG PFHK RFSCIIGPNG SGKSNV IDS MLFVFGYRAQ KIRSKKL SV ...String: MAHHHHHHHH HHLEVLFQGP RKGTQPSTA RRREEGPPPP S PDGASSDA EPEPPSGRTE SP ATAAETA SEELDNRSLE EIL NSIPPP PPPAMTNEAG APRL MITHI VNQNFKSYAG EKILG PFHK RFSCIIGPNG SGKSNV IDS MLFVFGYRAQ KIRSKKL SV LIHNSDEHKD IQSCTVEV H FQKIIDKEGD DYEVIPNSN FYVSRTACRD NTSVYHISGK KKTFKDVGN LLRSHGIDLD H NRFLILQG EVEQIAMMKP KG QTEHDEG MLEYLEDIIG CGR LNEPIK VLCRRVEILN EHRG EKLNR VKMVEKEKDA LEGEK NIAI EFLTLENEIF RKKNHV CQY YIYELQKRIA EMETQKE KI HEDTKEINEK SNILSNEM K AKNKDVKDTE KKLNKITKF IEENKEKFTQ LDLEDVQVRE KLKHATSKA KKLEKQLQKD K EKVEEFKS IPAKSNNIIN ET TTRNNAL EKEKEKEEKK LKE VMDSLK QETQGLQKEK ESRE KELMG FSKSVNEARS KMDVA QSEL DIYLSRHNTA VSQLTK AKE ALIAASETLK ERKAAIR DI EGKLPQTEQE LKEKEKEL Q KLTQEETNFK SLVHDLFQK VEEAKSSLAM NRSRGKVLDA IIQEKKSGR IPGIYGRLGD L GAIDEKYD VAISSCCHAL DY IVVDSID IAQECVNFLK RQN IGVATF IGLDKMAVWA KKMT EIQTP ENTPRLFDLV KVKDE KIRQ AFYFALRDTL VADNLD QAT RVAYQKDRRW RVVTLQG QI IEQSGTMTGG GSKVMKGR M GSSLVIEISE EEVNKMESQ LQNDSKKAMQ IQEQKVQLEE RVVKLRHSE REMRNTLEKF T ASIQRLIE QEEYLNVQVK EL EANVLAT APDKKKQKLL EEN VSAFKT EYDAVAEKAG KVEA EVKRL HNTIVEINNH KLKAQ QDKL DKINKQLDEC ASAITK AQV AIKTADRNLQ KAQDSVL RT EKEIKDTEKE VDDLTAEL K SLEDKAAEVV KNTNAAEES LPEIQKEHRN LLQELKVIQE NEHALQKDA LSIKLKLEQI D GHIAEHNS KIKYWHKEIS KI SLHPIED NPIEEISVLS PED LEAIKN PDSITNQIAL LEAR CHEMK PNLGAIAEYK KKEEL YLQR VAELDKITYE RDSFRQ AYE DLRKQRLNEF MAGFYII TN KLKENYQMLT LGGDAELE L VDSLDPFSEG IMFSVRPPK KSWKKIFNLS GGEKTLSSLA LVFALHHYK PTPLYFMDEI D AALDFKNV SIVAFYIYEQ TK NAQFIII SLRNNMFEIS DRL IGIYKT YNITKSVAVN PKEI ASKGL C |
-Macromolecule #3: CapH
| Macromolecule | Name: CapH / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGPPGPALPA TMNNSSSETR GHPHSASSP SERVFPMPLP R KAPLNIPG TPVLEDFPQN DD EKERLQR RRSRVFDLQF STD SPRLLA SPSSRSIDIS ATIP KFTNT QITEHYSTCI KLSTE NKIT TKNAFGLHLI DFMSEI LKQ KDTEPTNFKV AAGTLDA ST ...String: MGPPGPALPA TMNNSSSETR GHPHSASSP SERVFPMPLP R KAPLNIPG TPVLEDFPQN DD EKERLQR RRSRVFDLQF STD SPRLLA SPSSRSIDIS ATIP KFTNT QITEHYSTCI KLSTE NKIT TKNAFGLHLI DFMSEI LKQ KDTEPTNFKV AAGTLDA ST KIYAVRVDAV HADVYRVL G GLGKDAPSLE EVEGHVADG SATEMGTTKK AVKPKKKHLH RTIEQNINN LNVSEADRKC E IDPMFQKT AASFDECSTA GV FLSTLHC QDYRSELLFP SDV QTLSTG EPLELPELGC VEMT DLKAP LQQCAEDRQI CPSLA GFQF TQWDSETHNE SVSALV DKF KKNDQVFDIN AEVDESD CG DFPDGSLGDD FDANDEPD H TAVGDHEEFR SWKEPCQVQ SCQEEMISLG DGDIRTMCPL LSMKPGEYS YFSPRTMSMW A GPDHWRFR PRRKQDAPSQ SE NKKKSTK KDFEIDFEDD IDF DVYFRK TKAATILTKS TLEN QNWRA TTLPTDFNYN VDTLV QLHL KPGTRLLKMA QGHRVE TEH YEEIEDYDYN NPNDTSN FC PGLQAADSDD EDLDDLFV G PVGNSDLSPY PCHPPKTAQ QNGDTPEAQG LDITTYGESN LVAEPQKVN KIEIHYAKTA K KMDMKKLK QSMWSLLTAL SG KEADAEA NHREAGKEAA LAE VADEKM LSGLTKDLQR SLPP VMAQN LSIPLAFACL LHLAN EKNL KLEGTEDLSD VLVRQG DEN LYFQSWSHPQ FEKGGGS GG GSGGGSWSHP QFEK |
-Macromolecule #4: CapD2
| Macromolecule | Name: CapD2 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAPQMYEFHL PLSPEELLKS GGVNQYVVQ EVLSIKHLPP Q LRAFQAAF RAQGPLAMLQ HF DTIYSIL HHFRSIDPGL KED TLQFLI KVVSRHSQEL PAIL DDTTL SGSDRNAHLN ALKMN CYAL IRLLESFETM ASQTNL VDL DLGGKGKKAR TKAAHGF DW ...String: MAPQMYEFHL PLSPEELLKS GGVNQYVVQ EVLSIKHLPP Q LRAFQAAF RAQGPLAMLQ HF DTIYSIL HHFRSIDPGL KED TLQFLI KVVSRHSQEL PAIL DDTTL SGSDRNAHLN ALKMN CYAL IRLLESFETM ASQTNL VDL DLGGKGKKAR TKAAHGF DW EEERQPILQL LTQLLQLD I RHLWNHSIIE EEFVSLVTG CCYRLLENPT INHQKNRPTR EAITHLLGV ALTRYNHMLS A TVKIIQML QHFEHLAPVL VA AVSLWAT DYGMKSIVGE IVR EIGQKC PQELSRDPSG TKGF AAFLT ELAERVPAIL MSSMC ILLD HLDGENYMMR NAVLAA MAE MVLQVLSGDQ LEAAARD TR DQFLDTLQAH GHDVNSFV R SRVLQLFTRI VQQKALPLT RFQAVVALAV GRLADKSVLV CKNAIQLLA SFLANNPFSC K LSDADLAG PLQKETQKLQ EM RAQRRTA AASAVLDPEE EWE AMLPEL KSTLQQLLQL PQGE EEIPE QIANTETTED VKGRI YQLL AKASYKKAII LTREAT GHF QESEPFSHID PEESEET RL LNILGLIFKG PAASTQEK N PRESTGNMVT GQTVCKNKP NMSDPEESRG NDELVKQEML VQYLQDAYS FSRKITEAIG I ISKMMYEN TTTVVQEVIE FF VMVFQFG VPQALFGVRR MLP LIWSKE PGVREAVLNA YRQL YLNPK GDSARAKAQA LIQNL SLLL VDASVGTIQC LEEILC EFV QKDELKPAVT QLLWERA TE KVACCPLERC SSVMLLGM M ARGKPEIVGS NLDTLVSIG LDEKFPQDYR LAQQVCHAIA NISDRRKPS LGKRHPPFRL P QEHRLFER LRETVTKGFV HP DPLWIPF KEVAVTLIYQ LAE GPEVIC AQILQGCAKQ ALEK LEEKR TSQEDPKESP AMLPT FLLM NLLSLAGDVA LQQLVH LEQ AVSGELCRRR VLREEQE HK TKDPKEKNTS SETTMEEE L GLVGATADDT EAELIRGIC EMELLDGKQT LAAFVPLLLK VCNNPGLYS NPDLSAAASL A LGKFCMIS ATFCDSQLRL LF TMLEKSP LPIVRSNLMV ATG DLAIRF PNLVDPWTPH LYAR LRDPA QQVRKTAGLV MTHLI LKDM VKVKGQVSEM AVLLID PEP QIAALAKNFF NELSHKG NA IYNLLPDIIS RLSDPELG V EEEPFHTIMK QLLSYITKD KQTESLVEKL CQRFRTSRTE RQQRDLAYC VSQLPLTERG L RKMLDNFD CFGDKLSDES IF SAFLSVV GKLRRGAKPE GKA IIDEFE QKLRACHTRG LDGI KELEI GQAGSQRAPS AKKPS TGSR YQPLASTASD NDFVTP EPR RTTRRHPNTQ QRASKKK PK VVFSSDESSE EDLSAEMT E DETPKKTTPI LRASARRHR S |
-Macromolecule #5: CapG
| Macromolecule | Name: CapG / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGAERRLLSI KEAFRLAQQP HQNQAKLVV ALSRTYRTMD D KTVFHEEF IHYLKYVMVV YK REPAVER VIEFAAKFVT SFH QSDMED DEEEEDGGLL NYLF TFLLK SHEANSNAVR FRVCL LINK LLGSMPENAQ IDDDVF DKI NKAMLIRLKD KIPNVRI QA ...String: MGAERRLLSI KEAFRLAQQP HQNQAKLVV ALSRTYRTMD D KTVFHEEF IHYLKYVMVV YK REPAVER VIEFAAKFVT SFH QSDMED DEEEEDGGLL NYLF TFLLK SHEANSNAVR FRVCL LINK LLGSMPENAQ IDDDVF DKI NKAMLIRLKD KIPNVRI QA VLALSRLQDP KDDECPVV N AYATLIENDS NPEVRRAVL SCIAPSAKTL PKIVGRTKDV KEAVRKLAY QVLAEKVHMR A MSIAQRVM LLQQGLNDRS DA VKQAMQK HLLQGWLRFS EGN ILELLH RLDVENSSEV AVSV LNALF SITPLSELVG LCKNN DGRK LIPVETLTPE IALYWC ALC EYLKSKGDEG EEFLEQI LP EPVVYADYLL SYIQSIPV V NEEHRGDFSY IGNLMTKEF IGQQLILIIK SLDTSEEGGR KKLLAVLQE ILILPTIPIS L VSFLVERL LHIIIDDNKR TQ IVTEIIS EIRAPIVTVG VNN DPADVR KKELKMAEIK VKLI EAKEA LENCITLQDF NRASE LKEE IKALEDARIN LLKETE QLE IKEVHIEKND AETLQKC LI LCYELLKQMS ISTGLSAT M NGIIESLILP GIISIHPVV RNLAVLCLGC CGLQNQDFAR KHFVLLLQV LQIDDVTIKI S ALKAIFDQ LMTFGIEPFK TK KIKTLHC EGTEINSDDE QES KEVEET ATAKNVLKLL SDFL DSEVS ELRTGAAEGL AKLMF SGLL VSSRILSRLI LLWYNP VTE EDVQLRHCLG VFFPVFA YA SRTNQECFEE AFLPTLQT L ANAPASSPLA EIDITNVAE LLVDLTRPSG LNPQAKTSQD YQALTVHDN LAMKICNEIL T SPCSPEIR VYTKALSSLE LS SHLAKDL LVLLNEILEQ VKD RTCLRA LEKIKIQLEK GNKE FGDQA EAAQDATLTT TTFQN EDEK NKEVYMTPLR GVKATQ ASK STQLKTNRGQ RKVTVSA RT NRRCQTAEAD SESDHEVP E PESEMKMRLP RRAKTAALE KSKLNLAQFL NEDLS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: Grafix crosslinked sample was gel filtered on a superose 6 column into EM buffer (20 mM HEPES [pH 8], 200 mM KCl) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: The grids were glow discharged for 1 minute at 15 mA, the sample applied for 10 seconds and then washed twice with water before staining with 2% uranyl acetate for 1 minute before blotting and drying. | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 15.6 µm / Number grids imaged: 1 / Number real images: 862 / Average exposure time: 1.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)