+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10725 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | OP protein cage, complex 2 | |||||||||
 Map data Map data | OP protein cage, complex 2 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Salmonella enterica (bacteria) Salmonella enterica (bacteria) | |||||||||
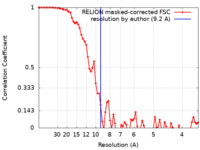
| Method | single particle reconstruction / cryo EM / Resolution: 9.2 Å | |||||||||
 Authors Authors | Edwardson TGW / Tetter S / Hilvert D | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Two-tier supramolecular encapsulation of small molecules in a protein cage. Authors: Thomas G W Edwardson / Stephan Tetter / Donald Hilvert /  Abstract: Expanding protein design to include other molecular building blocks has the potential to increase structural complexity and practical utility. Nature often employs hybrid systems, such as clathrin- ...Expanding protein design to include other molecular building blocks has the potential to increase structural complexity and practical utility. Nature often employs hybrid systems, such as clathrin-coated vesicles, lipid droplets, and lipoproteins, which combine biopolymers and lipids to transport a broader range of cargo molecules. To recapitulate the structure and function of such composite compartments, we devised a supramolecular strategy that enables porous protein cages to encapsulate poorly water-soluble small molecule cargo through templated formation of a hydrophobic surfactant-based core. These lipoprotein-like complexes protect their cargo from sequestration by serum proteins and enhance the cellular uptake of fluorescent probes and cytotoxic drugs. This design concept could be applied to other protein cages, surfactant mixtures, and cargo molecules to generate unique hybrid architectures and functional capabilities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10725.map.gz emd_10725.map.gz | 27.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10725-v30.xml emd-10725-v30.xml emd-10725.xml emd-10725.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10725_fsc.xml emd_10725_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10725.png emd_10725.png | 114.4 KB | ||
| Masks |  emd_10725_msk_1.map emd_10725_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_10725_half_map_1.map.gz emd_10725_half_map_1.map.gz emd_10725_half_map_2.map.gz emd_10725_half_map_2.map.gz | 20.9 MB 20.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10725 http://ftp.pdbj.org/pub/emdb/structures/EMD-10725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10725 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10725.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10725.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OP protein cage, complex 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
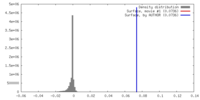
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
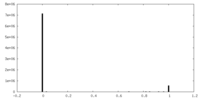
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10725_msk_1.map emd_10725_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
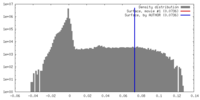
| Density Histograms |
-Half map: #2
| File | emd_10725_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
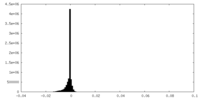
| Density Histograms |
-Half map: #1
| File | emd_10725_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OP protein cage
| Entire | Name: OP protein cage |
|---|---|
| Components |
|
-Supramolecule #1: OP protein cage
| Supramolecule | Name: OP protein cage / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Positively supercharged variant of the computationally designed cage protein O3-33 |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: OP protein cage, complex 2
| Macromolecule | Name: OP protein cage, complex 2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MSQAIGILEL RSIAAGMELG DAMLKSANVD LLVSKTISRG KFLLMLGGDI GAIQQAIETG TSQAGRLLVD SLVLANIHPS VLPAISGLNS VDKRQAVGIV ETRSVAACIS AADRAVKGSN VTLVRVHMAR GIGGKCYMVV AGDVSDVALA VTVASSSAGA YGRLVYASLI PRPHEAMWRQ MVEGLEHHHH HH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 25 mM Tris-HCl, 200 mM NaCl, 5 mM EDTA, pH 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 22 K / Details: blot for 12 s, 25 blot strength, before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-7 / Number grids imaged: 1 / Number real images: 127 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: -3.3 µm / Nominal defocus min: -1.8 µm / Nominal magnification: 62000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)