+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10625 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cin8 motor domain (short loop 8) bound to microtubules | |||||||||
 Map data Map data | Cin8 motor domain (short loop 8) bound to microtubules | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinesin / Cin8 / Motor domain / Microtubules / MOTOR PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 6.1 Å | |||||||||
 Authors Authors | Siegler N / Singh SK / Zalk R / Debs GE / Sindelar CV / Gheber L / Zarivach R | |||||||||
| Funding support |  Israel, 1 items Israel, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Noncanonical interaction with microtubules via the N-terminal nonmotor domain is critical for the functions of a bidirectional kinesin. Authors: Sudhir K Singh / Nurit Siegler / Himanshu Pandey / Neta Yanir / Mary Popov / Alina Goldstein-Levitin / Mayan Sadan / Garrett Debs / Raz Zarivach / Gabriel A Frank / Itamar Kass / Charles V ...Authors: Sudhir K Singh / Nurit Siegler / Himanshu Pandey / Neta Yanir / Mary Popov / Alina Goldstein-Levitin / Mayan Sadan / Garrett Debs / Raz Zarivach / Gabriel A Frank / Itamar Kass / Charles V Sindelar / Ran Zalk / Larisa Gheber /   Abstract: Several kinesin-5 motors (kinesin-5s) exhibit bidirectional motility. The mechanism of such motility remains unknown. Bidirectional kinesin-5s share a long N-terminal nonmotor domain (NTnmd), absent ...Several kinesin-5 motors (kinesin-5s) exhibit bidirectional motility. The mechanism of such motility remains unknown. Bidirectional kinesin-5s share a long N-terminal nonmotor domain (NTnmd), absent in exclusively plus-end-directed kinesins. Here, we combined in vivo, in vitro, and cryo-electron microscopy (cryo-EM) studies to examine the impact of NTnmd mutations on the motor functions of the bidirectional kinesin-5, Cin8. We found that NTnmd deletion mutants exhibited cell viability and spindle localization defects. Using cryo-EM, we examined the structure of a microtubule (MT)-bound motor domain of Cin8, containing part of its NTnmd. Modeling and molecular dynamic simulations based on the cryo-EM map suggested that the NTnmd of Cin8 interacts with the C-terminal tail of β-tubulin. In vitro experiments on subtilisin-treated MTs confirmed this notion. Last, we showed that NTnmd mutants are defective in plus-end-directed motility in single-molecule and antiparallel MT sliding assays. These findings demonstrate that the NTnmd, common to bidirectional kinesin-5s, is critical for their bidirectional motility and intracellular functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10625.map.gz emd_10625.map.gz | 51.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10625-v30.xml emd-10625-v30.xml emd-10625.xml emd-10625.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10625.png emd_10625.png | 132.5 KB | ||
| Filedesc metadata |  emd-10625.cif.gz emd-10625.cif.gz | 5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10625 http://ftp.pdbj.org/pub/emdb/structures/EMD-10625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10625 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10625.map.gz / Format: CCP4 / Size: 58.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10625.map.gz / Format: CCP4 / Size: 58.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cin8 motor domain (short loop 8) bound to microtubules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
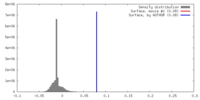
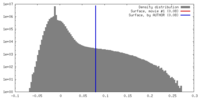
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ALPHA and Beta TUBULIN WITH CIN8 MOTOR DOMAIN
| Entire | Name: ALPHA and Beta TUBULIN WITH CIN8 MOTOR DOMAIN |
|---|---|
| Components |
|
-Supramolecule #1: ALPHA and Beta TUBULIN WITH CIN8 MOTOR DOMAIN
| Supramolecule | Name: ALPHA and Beta TUBULIN WITH CIN8 MOTOR DOMAIN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: ADDED 0.1 mM AMP-PNP, 10 micromolar Taxol |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cin8, microtubules
| Macromolecule | Name: Cin8, microtubules / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MPAENQNTGQ DRSSNSISKN GNSQVGCHTV PNEELNITVA VRCRGRNERE ISMKSSVVVN VPDITGSKEI SINTTGDTGI TAQMNAKRYT VDKVFGPGAS QDLIFDEVAG PLFQDFIKGY NCTVLVYGMT STGKTYTMTG DEKLYNGELS DAAGIIPRVL LKLFDTLELQ ...String: MPAENQNTGQ DRSSNSISKN GNSQVGCHTV PNEELNITVA VRCRGRNERE ISMKSSVVVN VPDITGSKEI SINTTGDTGI TAQMNAKRYT VDKVFGPGAS QDLIFDEVAG PLFQDFIKGY NCTVLVYGMT STGKTYTMTG DEKLYNGELS DAAGIIPRVL LKLFDTLELQ QNDYVVKCSF IELYNEELKD LLDSNSNGSS NTGFDGQFMK KLRIFDNNNN NSSIYIQNLQ EFHITNAMEG LNLLQKGLKH RQVASTKMND FSSRSHTIFT ITLYKKHQDE LFRISKMNLV DLAGSENINR SGALNQRAKE AGSINQSLLT LGRVINALVD KSGHIPFRES KLTRLLQDSL GGNTKTALIA TISPAKVTSE ETCSTLEYAS KAKNIKNKPQ LGSFIMKDIL LEHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.9 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.3 mm |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 82.18 Å Applied symmetry - Helical parameters - Δ&Phi: -0.53 ° Applied symmetry - Helical parameters - Axial symmetry: C14 (14 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 6.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 47551 |
|---|---|
| Segment selection | Number selected: 47551 / Software - Name: RELION (ver. 2.1) / Details: Number of particles seleced for reconstruction |
| Startup model | Type of model: INSILICO MODEL |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION (ver. 2.1) |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)