[English] 日本語
 Yorodumi
Yorodumi- EMDB-10578: Ubiquitin Ligation to substrate by a cullin-RING E3 ligase: NEDD8... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10578 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase: NEDD8-CUL1-RBX1 SKP1-dimeric bTRCP2-IkBa-(UB)~UBE2D2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
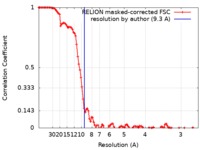
| Method | single particle reconstruction / cryo EM / Resolution: 9.3 Å | |||||||||
 Authors Authors | Baek K / Prabu JR / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Authors: Kheewoong Baek / David T Krist / J Rajan Prabu / Spencer Hill / Maren Klügel / Lisa-Marie Neumaier / Susanne von Gronau / Gary Kleiger / Brenda A Schulman /   Abstract: Eukaryotic cell biology depends on cullin-RING E3 ligase (CRL)-catalysed protein ubiquitylation, which is tightly controlled by the modification of cullin with the ubiquitin-like protein NEDD8. ...Eukaryotic cell biology depends on cullin-RING E3 ligase (CRL)-catalysed protein ubiquitylation, which is tightly controlled by the modification of cullin with the ubiquitin-like protein NEDD8. However, how CRLs catalyse ubiquitylation, and the basis of NEDD8 activation, remain unknown. Here we report the cryo-electron microscopy structure of a chemically trapped complex that represents the ubiquitylation intermediate, in which the neddylated CRL1 promotes the transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme UBE2D to its recruited substrate, phosphorylated IκBα. NEDD8 acts as a nexus that binds disparate cullin elements and the RING-activated ubiquitin-linked UBE2D. Local structural remodelling of NEDD8 and large-scale movements of CRL domains converge to juxtapose the substrate and the ubiquitylation active site. These findings explain how a distinctive ubiquitin-like protein alters the functions of its targets, and show how numerous NEDD8-dependent interprotein interactions and conformational changes synergistically configure a catalytic CRL architecture that is both robust, to enable rapid ubiquitylation of the substrate, and fragile, to enable the subsequent functions of cullin-RING proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10578.map.gz emd_10578.map.gz | 6.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10578-v30.xml emd-10578-v30.xml emd-10578.xml emd-10578.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10578_fsc.xml emd_10578_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10578.png emd_10578.png | 48.3 KB | ||
| Masks |  emd_10578_msk_1.map emd_10578_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_10578_half_map_1.map.gz emd_10578_half_map_1.map.gz emd_10578_half_map_2.map.gz emd_10578_half_map_2.map.gz | 79.3 MB 79.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10578 http://ftp.pdbj.org/pub/emdb/structures/EMD-10578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10578 | HTTPS FTP |
-Validation report
| Summary document |  emd_10578_validation.pdf.gz emd_10578_validation.pdf.gz | 387.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10578_full_validation.pdf.gz emd_10578_full_validation.pdf.gz | 387.1 KB | Display | |
| Data in XML |  emd_10578_validation.xml.gz emd_10578_validation.xml.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10578 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10578 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10578 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10578 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10578.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10578.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10578_msk_1.map emd_10578_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10578_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10578_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ubiquitin Ligation to substrate by a cullin-RING E3 ligase: NEDD8...
| Entire | Name: Ubiquitin Ligation to substrate by a cullin-RING E3 ligase: NEDD8-CUL1-RBX1 SKP1-dimeric bTRCP2-IkB-(UB)~UBE2D2 |
|---|---|
| Components |
|
-Supramolecule #1: Ubiquitin Ligation to substrate by a cullin-RING E3 ligase: NEDD8...
| Supramolecule | Name: Ubiquitin Ligation to substrate by a cullin-RING E3 ligase: NEDD8-CUL1-RBX1 SKP1-dimeric bTRCP2-IkB-(UB)~UBE2D2 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 210 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)