[English] 日本語
 Yorodumi
Yorodumi- EMDB-10552: Helical reconstruction of AdhE from Escherichia coli in its exten... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10552 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of AdhE from Escherichia coli in its extended conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
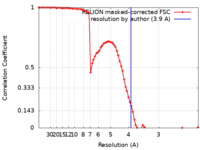
| Method | helical reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Fronzes R / Pony P | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation. Authors: Pauline Pony / Chiara Rapisarda / Laurent Terradot / Esther Marza / Rémi Fronzes /   Abstract: Acetaldehyde-alcohol dehydrogenase (AdhE) enzymes are a key metabolic enzyme in bacterial physiology and pathogenicity. They convert acetyl-CoA to ethanol via an acetaldehyde intermediate during ...Acetaldehyde-alcohol dehydrogenase (AdhE) enzymes are a key metabolic enzyme in bacterial physiology and pathogenicity. They convert acetyl-CoA to ethanol via an acetaldehyde intermediate during ethanol fermentation in an anaerobic environment. This two-step reaction is associated to NAD regeneration, essential for glycolysis. The bifunctional AdhE enzyme is conserved in all bacterial kingdoms but also in more phylogenetically distant microorganisms such as green microalgae. It is found as an oligomeric form called spirosomes, for which the function remains elusive. Here, we use cryo-electron microscopy to obtain structures of Escherichia coli spirosomes in different conformational states. We show that spirosomes contain active AdhE monomers, and that AdhE filamentation is essential for its activity in vitro and function in vivo. The detailed analysis of these structures provides insight showing that AdhE filamentation is essential for substrate channeling within the filament and for the regulation of enzyme activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10552.map.gz emd_10552.map.gz | 31 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10552-v30.xml emd-10552-v30.xml emd-10552.xml emd-10552.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10552_fsc.xml emd_10552_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10552.png emd_10552.png | 52.8 KB | ||
| Others |  emd_10552_additional.map.gz emd_10552_additional.map.gz emd_10552_additional_1.map.gz emd_10552_additional_1.map.gz emd_10552_half_map_1.map.gz emd_10552_half_map_1.map.gz emd_10552_half_map_2.map.gz emd_10552_half_map_2.map.gz | 10.5 MB 10.5 MB 31.3 MB 31.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10552 http://ftp.pdbj.org/pub/emdb/structures/EMD-10552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10552 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10552.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10552.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
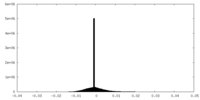
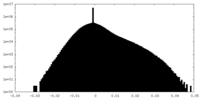
-Additional map: #1
| File | emd_10552_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_10552_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
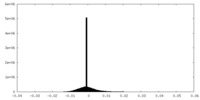
-Half map: #1
| File | emd_10552_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
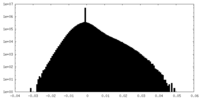
-Half map: #2
| File | emd_10552_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AdhE dimer in complex with NAD+ and Fe2+
| Entire | Name: AdhE dimer in complex with NAD+ and Fe2+ |
|---|---|
| Components |
|
-Supramolecule #1: AdhE dimer in complex with NAD+ and Fe2+
| Supramolecule | Name: AdhE dimer in complex with NAD+ and Fe2+ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 0.0025 µm / Calibrated defocus min: 0.0005 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)