[English] 日本語
 Yorodumi
Yorodumi- EMDB-10537: Yeast 80S ribosome in complex with eIF5A and decoding A-site and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10537 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | |||||||||
 Map data Map data | Cryo-EM density of the yeast 80S ribosome in complex with eIF5A and A-site and P-site tRNAs. Filtered according to local resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | eIF5A / translation control / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoplasmic translational elongation through polyproline stretches / positive regulation of cytoplasmic translational elongation through polyproline stretches / Hypusine synthesis from eIF5A-lysine / CAT tailing / translational frameshifting / cytoplasmic translational elongation / positive regulation of translational termination / cytoplasmic translational termination / positive regulation of translational elongation / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, LSU-rRNA,5S) ...cytoplasmic translational elongation through polyproline stretches / positive regulation of cytoplasmic translational elongation through polyproline stretches / Hypusine synthesis from eIF5A-lysine / CAT tailing / translational frameshifting / cytoplasmic translational elongation / positive regulation of translational termination / cytoplasmic translational termination / positive regulation of translational elongation / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, LSU-rRNA,5S) / regulation of amino acid metabolic process / negative regulation of glucose mediated signaling pathway / positive regulation of translational fidelity / RMTs methylate histone arginines / Protein methylation / mTORC1-mediated signalling / Protein hydroxylation / ribosome-associated ubiquitin-dependent protein catabolic process / GDP-dissociation inhibitor activity / positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / pre-mRNA 5'-splice site binding / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / Ribosomal scanning and start codon recognition / preribosome, small subunit precursor / nonfunctional rRNA decay / response to cycloheximide / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Major pathway of rRNA processing in the nucleolus and cytosol / mRNA destabilization / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / negative regulation of translational frameshifting / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / negative regulation of mRNA splicing, via spliceosome / Formation of a pool of free 40S subunits / preribosome, large subunit precursor / positive regulation of protein kinase activity / L13a-mediated translational silencing of Ceruloplasmin expression / positive regulation of translational initiation / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal large subunit export from nucleus / translational elongation / G-protein alpha-subunit binding / 90S preribosome / translation elongation factor activity / ribosomal subunit export from nucleus / regulation of translational fidelity / protein-RNA complex assembly / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translational termination / maturation of LSU-rRNA / ribosomal small subunit export from nucleus / translation regulator activity / DNA-(apurinic or apyrimidinic site) endonuclease activity / translation initiation factor activity / rescue of stalled ribosome / cellular response to amino acid starvation / protein kinase C binding / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal large subunit biogenesis / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosome assembly / maturation of SSU-rRNA / small-subunit processome / translational initiation / macroautophagy / maintenance of translational fidelity / modification-dependent protein catabolic process / protein tag activity / cytoplasmic stress granule / rRNA processing / ribosome biogenesis / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / small ribosomal subunit rRNA binding / ribosomal large subunit assembly / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / rRNA binding / protein ubiquitination / structural constituent of ribosome / ribosome / translation / G protein-coupled receptor signaling pathway / ribonucleoprotein complex / negative regulation of gene expression / response to antibiotic / mRNA binding / ubiquitin protein ligase binding / nucleolus / perinuclear region of cytoplasm / mitochondrion Similarity search - Function | |||||||||
| Biological species |  | |||||||||
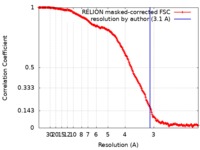
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Buschauer R / Cheng J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: The Ccr4-Not complex monitors the translating ribosome for codon optimality. Authors: Robert Buschauer / Yoshitaka Matsuo / Takato Sugiyama / Ying-Hsin Chen / Najwa Alhusaini / Thomas Sweet / Ken Ikeuchi / Jingdong Cheng / Yasuko Matsuki / Risa Nobuta / Andrea Gilmozzi / Otto ...Authors: Robert Buschauer / Yoshitaka Matsuo / Takato Sugiyama / Ying-Hsin Chen / Najwa Alhusaini / Thomas Sweet / Ken Ikeuchi / Jingdong Cheng / Yasuko Matsuki / Risa Nobuta / Andrea Gilmozzi / Otto Berninghausen / Petr Tesina / Thomas Becker / Jeff Coller / Toshifumi Inada / Roland Beckmann /    Abstract: Control of messenger RNA (mRNA) decay rate is intimately connected to translation elongation, but the spatial coordination of these events is poorly understood. The Ccr4-Not complex initiates mRNA ...Control of messenger RNA (mRNA) decay rate is intimately connected to translation elongation, but the spatial coordination of these events is poorly understood. The Ccr4-Not complex initiates mRNA decay through deadenylation and activation of decapping. We used a combination of cryo-electron microscopy, ribosome profiling, and mRNA stability assays to examine the recruitment of Ccr4-Not to the ribosome via specific interaction of the Not5 subunit with the ribosomal E-site in This interaction occurred when the ribosome lacked accommodated A-site transfer RNA, indicative of low codon optimality. Loss of the interaction resulted in the inability of the mRNA degradation machinery to sense codon optimality. Our findings elucidate a physical link between the Ccr4-Not complex and the ribosome and provide mechanistic insight into the coupling of decoding efficiency with mRNA stability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10537.map.gz emd_10537.map.gz | 235.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10537-v30.xml emd-10537-v30.xml emd-10537.xml emd-10537.xml | 95.7 KB 95.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10537_fsc.xml emd_10537_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10537.png emd_10537.png | 89.7 KB | ||
| Filedesc metadata |  emd-10537.cif.gz emd-10537.cif.gz | 18.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10537 http://ftp.pdbj.org/pub/emdb/structures/EMD-10537 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10537 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10537 | HTTPS FTP |
-Validation report
| Summary document |  emd_10537_validation.pdf.gz emd_10537_validation.pdf.gz | 512.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10537_full_validation.pdf.gz emd_10537_full_validation.pdf.gz | 512 KB | Display | |
| Data in XML |  emd_10537_validation.xml.gz emd_10537_validation.xml.gz | 15.6 KB | Display | |
| Data in CIF |  emd_10537_validation.cif.gz emd_10537_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10537 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10537 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10537 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10537 | HTTPS FTP |
-Related structure data
| Related structure data |  6tnuMC  6tb3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10537.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10537.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM density of the yeast 80S ribosome in complex with eIF5A and A-site and P-site tRNAs. Filtered according to local resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Yeast 80S ribosome in complex with eIF5A programed by A-site and ...
+Supramolecule #1: Yeast 80S ribosome in complex with eIF5A programed by A-site and ...
+Macromolecule #1: 18S rRNA
+Macromolecule #2: mRNA
+Macromolecule #24: P-site tRNA
+Macromolecule #37: A-site tRNA
+Macromolecule #38: 25S rRNA
+Macromolecule #39: 5S rRNA
+Macromolecule #40: 5.8S rRNA
+Macromolecule #3: 40S ribosomal protein S0-A
+Macromolecule #4: 40S ribosomal protein S1-A
+Macromolecule #5: 40S ribosomal protein S15
+Macromolecule #6: 40S ribosomal protein S2
+Macromolecule #7: 40S ribosomal protein S3
+Macromolecule #8: 40S ribosomal protein S4-A
+Macromolecule #9: Rps5p
+Macromolecule #10: 40S ribosomal protein S6-A
+Macromolecule #11: 40S ribosomal protein S7-A
+Macromolecule #12: 40S ribosomal protein S8
+Macromolecule #13: 40S ribosomal protein S9-A
+Macromolecule #14: 40S ribosomal protein S10-A
+Macromolecule #15: 40S ribosomal protein S11-A
+Macromolecule #16: 40S ribosomal protein S12
+Macromolecule #17: 40S ribosomal protein S13
+Macromolecule #18: 40S ribosomal protein S14-B
+Macromolecule #19: 40S ribosomal protein S16-A
+Macromolecule #20: 40S ribosomal protein S17-B
+Macromolecule #21: 40S ribosomal protein S18-A
+Macromolecule #22: 40S ribosomal protein S19-A
+Macromolecule #23: 40S ribosomal protein S20
+Macromolecule #25: 40S ribosomal protein S21-A
+Macromolecule #26: 40S ribosomal protein S22-A
+Macromolecule #27: 40S ribosomal protein S23-A
+Macromolecule #28: 40S ribosomal protein S24-A
+Macromolecule #29: 40S ribosomal protein S25-A
+Macromolecule #30: 40S ribosomal protein S26-B
+Macromolecule #31: 40S ribosomal protein S27-A
+Macromolecule #32: 40S ribosomal protein S29-A
+Macromolecule #33: 40S ribosomal protein S30-A
+Macromolecule #34: Ubiquitin-40S ribosomal protein S31
+Macromolecule #35: Guanine nucleotide-binding protein subunit beta-like protein
+Macromolecule #36: 40S ribosomal protein S28-A
+Macromolecule #41: 60S ribosomal protein L2-A
+Macromolecule #42: 60S ribosomal protein L3
+Macromolecule #43: 60S ribosomal protein L4-A
+Macromolecule #44: 60S ribosomal protein L5
+Macromolecule #45: 60S ribosomal protein L6-B
+Macromolecule #46: 60S ribosomal protein L7-A
+Macromolecule #47: 60S ribosomal protein L8-A
+Macromolecule #48: 60S ribosomal protein L9-A
+Macromolecule #49: 60S ribosomal protein L10
+Macromolecule #50: 60S ribosomal protein L11-B
+Macromolecule #51: 60S ribosomal protein L13-A
+Macromolecule #52: 60S ribosomal protein L14-A
+Macromolecule #53: 60S ribosomal protein L15-A
+Macromolecule #54: 60S ribosomal protein L16-A
+Macromolecule #55: 60S ribosomal protein L17-A
+Macromolecule #56: 60S ribosomal protein L18-A
+Macromolecule #57: 60S ribosomal protein L19-A
+Macromolecule #58: 60S ribosomal protein L20-A
+Macromolecule #59: 60S ribosomal protein L21-A
+Macromolecule #60: 60S ribosomal protein L22-A
+Macromolecule #61: 60S ribosomal protein L23-A
+Macromolecule #62: 60S ribosomal protein L24-A
+Macromolecule #63: 60S ribosomal protein L25
+Macromolecule #64: 60S ribosomal protein L26-A
+Macromolecule #65: 60S ribosomal protein L27-A
+Macromolecule #66: 60S ribosomal protein L28
+Macromolecule #67: 60S ribosomal protein L29
+Macromolecule #68: 60S ribosomal protein L30
+Macromolecule #69: 60S ribosomal protein L31-A
+Macromolecule #70: 60S ribosomal protein L32
+Macromolecule #71: 60S ribosomal protein L33-A
+Macromolecule #72: 60S ribosomal protein L34-A
+Macromolecule #73: 60S ribosomal protein L35-A
+Macromolecule #74: 60S ribosomal protein L36-A
+Macromolecule #75: 60S ribosomal protein L37-A
+Macromolecule #76: 60S ribosomal protein L38
+Macromolecule #77: 60S ribosomal protein L39
+Macromolecule #78: Ubiquitin-60S ribosomal protein L40
+Macromolecule #79: 60S ribosomal protein L41-A
+Macromolecule #80: 60S ribosomal protein L42-A
+Macromolecule #81: 60S ribosomal protein L43-A
+Macromolecule #82: Eukaryotic translation initiation factor 5A-1
+Macromolecule #83: 60S ribosomal protein L1
+Macromolecule #84: ZINC ION
+Macromolecule #85: SPERMIDINE
+Macromolecule #86: 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethy...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)